Development of SARS-CoV-2 Isothermal Amplification Detection Kits
Xueling Li 1, Jing Tian 1, Yan Xu 1, Xiaomin Chen 1, 2, Zexi Liu 1, Hui Liang 1, Caihong Jin 1, Daxiang Cui 1, 2 *
1 National Engineering Research Center for Nanotechnology, Shanghai, China.
2 Department of Instrument Science and Engineering, Shanghai Jiao Tong University, Shanghai, China.
* Corresponding author. E-mail: dxcui@sjtu.edu.cn
Received: Aug. 24, 2020; Accepted: Oct. 28, 2020; Published: Oct. 28, 2020
Citation: Xueling Li, Jing Tian, Yan Xu, Xiaomin Chen, Zexi Liu, Hui Liang, Caihong Jin, and Daxiang Cui, Development of SARS-CoV-2 Isothermal Amplification Detection Kits. Nano Biomed. Eng., 2020, 12(4): 316-320.
DOI: 10.5101/nbe.v12i4.p316-320.
Abstract
The SARS-CoV-2 isothermal amplification detection kits based on loop-mediated isothermal amplification (LAMP) were developed and evaluated on three types samples of SARS-CoV-2. The kits included enzyme reaction mixtures and chromogenic agents. After the isothermal amplification reactions were completed, the reaction results were judged by using the chromogenic agents to determine whether SARS-CoV-2 exists in the samples to be tested. The detection kits have the advantages of convenient operation, fast detection speed and high sensitivity up to 1 copy of virus particles per reaction, which can speed up the detection speed of suspected cases, and avoid the missing detection problems caused by the low detection sensitivity.
Keywords: COVID-19, SARS-CoV-2, Detection kits, Isothermal amplification, Loop-mediated isothermal amplification
From December 2019, COVID-19 caused by SARS-CoV-2 (2019-nCoV) has become a public health event of global concern [1-3]. According to the latest data reported to the World Health Organization on 19 August 2020, there have been nearly 22 million confirmed cases and more than 770000 deaths caused by SARS-CoV-2 worldwide [4]. Currently, the importance of nucleic acid detection as a diagnostic criterion was generally recognized [3, 5]. However, nucleic acid detection based on real time PCR (RT-PCR) requires the fluorescence quantitative PCR instruments and a high level of operations [3, 6-11]. Therefore, the popularization and field application of nucleic acid detection are limited and a large number of suspected cases can’t be diagnosed in time. On the other hand, because the sensitivity of the current RT-PCR detection methods is not enough, especially in the early stage of disease course, it is easy to cause false-negative resulting in missed detection problems [6-10]. Hence, it is necessary to develop the detection mixtures/kits of SARS-CoV-2 with the advantages of convenient operation, short detection time and high sensitivity. Loop-mediated isothermal amplification (LAMP) method is expected to solve the current problems encountered in molecular detection of SARS-CoV-2 [12-17]. LAMP is a technology that provides nucleic acid amplification within 60 minutes under a constant temperature [12]. The amplification reaction can be performed on a simple device (e.g. water baths and electric heating devices) in addition to PCR instruments or quantitative PCR instruments and the results of reaction can be judged with the naked eyes or simple ultraviolet devices [13]. LAMP-based methods were developed for the detection of SARS-CoV-2 by several groups [12-17], e.g. the method based on the primer set NA developed by Zhang et al. can detect as low as 120 copies of SARS-CoV-2 per reaction [17]. In this paper, we reported the preliminary evaluation results of the kits prepared based on the optimized reaction mixtures with the above primer set NA. After optimizing, up to 1 copy of virus particles per reaction could be detected. The improvement of detection sensitivity will further reduce false-negative rate and avoid missed detection of COVID-19 to the maximum extent.
Experimental
The optimization of LAMP assay conditions
The enzyme reaction mixture using a combination of Bst DNA polymerase with thermopol buffer and Mg2+ ion (New England Biolab, Whitby, ON), dNTP (New England Biolab, Whitby, ON), betaine (Sangon Biotech (Shanghai) Co., Ltd.), reverse transcriptase (Sangon Biotech, Shanghai) and primers. The primer set NA used as Zhang et al. [15] was synthetized by Sangon Biotech (Shanghai) Co., Ltd. The concentration of reagent ingredients and reaction conditions were optimized according to the results of sensitivity tests. The chromogenic agents were used to judge the results of amplification reaction including Sybr Green I (10000×, Invitrogen, Burlington, ON, Canada), GeneFinder (10000×, Xiamen Zeesan Biotech, Fujian, China) and GelRed (10000×, Biotium, California, USA).
LAMP assays on RNA reference materials
SARS-CoV-2 RNA transcribed in vitro reference materials including the total length of nucleocapsid protein N gene, the total length of envelope protein E gene, open reading frame 1ab gene fragment, numbered GBE (E) 091112, with N gene concentration of 2.9×107 copies/μL, was provided by Shanghai Institute of Measurement and Testing Technology. RNA templates were added to the reaction mixture (Table 1) prepared by using the SARS-CoV-2 detection kits with 2×104, 2×103, 2×102, 20, 5, 2, 1 and 0 copies,respectively. Then isothermal amplifications were performed under 63 oC conditions for 40 minutes and terminated under 82 oC for 7 min. Then 50× of the final concentration of Sybr Green I, GeneFinder and GelRed were added to the amplification products respectively, to determine the sensitivity of the detection kits with the naked eyes, or the ultraviolet light at 254 nm of wavelength.
LAMP assays on RNA samples from pseudo-virus
SARS-CoV-2 pseudo-virus samples including the total length of nucleocapsid protein N gene (Catalogue No. FNV2565, concentration 108 copies/ml), were purchased from Shanghai Fubio Biotechnology Co. Ltd. The pseudo-virus samples were diluted in series with water, and then RNA was extracted from 200 μL of each diluted sample with TaKaRa nucleic acid extraction kit (Catalogue No. 9766) according to the instructions and eluted with 50 μL of water. The reaction mixtures were prepared according to the optimized conditions (Table 1) by using the SARS-CoV-2 detection kits, and then 5 μL of RNA extracted from the above measurement audit samples with TaKaRa nucleic acid extraction kit (Catalogue No. 9766) were added to the reaction mixtures with 104, 103, 102, 10, 2, 1 and 0 copies,respectively. After the isothermal amplification, GeneFinder was used as the chromogenic agent to the amplification products to determine the reaction results with the naked eyes and the ultraviolet light at 254 nm of wavelength.
LAMP assays on RNA samples from COVID-19 patients
Four SARS-CoV-2 nucleic acid detection measurement audit samples, numbered 2013, 2014, 2015, 2016 respectively, prepared from the inactivated clinical samples, were provided by the Shanghai Center for Clinical Laboratory. The sample No. 2013 was negative sample and the other three samples No. 2014, 2015 and 2016 were positive samples, of which No. 2016 was the sample with low concentration. The reaction mixtures were prepared according to the optimized conditions (Table 1) using the SARS-CoV-2 detection kits, and then 5 μL of RNA extracted from the above measurement audit samples with TaKaRa nucleic acid extraction kit (Catalogue No. 9766) were added to the reaction mixtures. The RNA reference materials provided by Shanghai Institute of Measurement and Testing Technology, and the DEPC-treated water were used as the positive control and the negative control, respectively. After the amplification, Sybr Green I was used as the chromogenic agent into the amplification products to determine the detection limits with the naked eyes.
Results and Discussion
The optimization of LAMP assay conditions
The optimized reaction was performed in a 25 μL reaction mixture containing 1×Bst DNA Polymerase reaction buffer, 6.0 mmol/L of Mg2+, 0.6 mol/L of betaine, 1.2 mmol/L of dNTP, 0.2 μmol/L of F3 and B3 primers, 1.6 μmol/L FIP and BIP primers, 0.4 μmol/L of LF and LB primers, 0.32 U/μL of Bst DNA polymerase, 0.06 U/μL of reverse transcriptase and 5 μL of RNA template (Table 1). And 1×Bst DNA polymerase reaction buffer contains 20 mmol/L of Tris-HCl (pH 8.8), 10 mmol/L of KCl, 10 mmol/L of (NH4)2SO4, 0.1% of Triton X-100 and 2 mM of MgSO4. The isothermal amplification reactions are performed at 63 ℃ for 40 minutes and then the enzyme was inactivated at 82 ℃ for 7 minutes. The isothermal amplification reaction can be set on a general PCR instrument, a fluorescence quantitative PCR instrument, a water bath, an electric heater and other temperature control devices. Sybr Green I, GeneFinder or GelRed are selected as the chromogenic agents with 40~50 fold of the working concentration. When Sybr Green I or GeneFinder is used as the chromogenic agent, the reaction results can be determined with the naked eyes or ultraviolet light at 254 nm or 365 nm wavelength. When GelRed is used as the chromogenic agent, the positive and negative results can’t be distinguished with the naked eyes, and it needs to be distinguished by ultraviolet light at 254 nm or 365 nm wavelength. For the kit, the components may be independent or premixed. In addition to positive, negative controls and the chromogenic agent, the kit comprises two or three premixed reagents. For example, the premixed reagent I comprises Bst DNA polymerase reaction buffer, Mg2+, betaine and dNTP and the premixed reagent II comprises the primer set (F3、B3、FIP、BIP、LF and LB) and premixed reagent III comprises Bst DNA polymerase and reverse transcriptase. The components within reagent II or III can also be mixed into premixed reagent I.
Table 1 Reaction mixture of SARS-CoV-2 detection kits
|
Reagent ingredients |
Working concentration |
|
Reaction buffer |
1×Bst DNA polymerase reaction buffer |
|
Mg2+ |
6.0 mmol/L |
|
Betain |
0.6 mol/L |
|
dNTP |
1.2 mmol/L |
|
Primer F3 |
0.2 µmol/L |
|
Primer B3 |
0.2 µmol/L |
|
Primer FIP |
1.6 µmol/L |
|
Primer BIP |
1.6 µmol/L |
|
Primer LF |
0.4 µmol/L |
|
Primer LB |
0.4 µmol/L |
|
Reverse transcriptase |
0.06 U/ μL |
|
Bst DNA polymerase |
0.32 U/μL |
LAMP assays on RNA reference materials
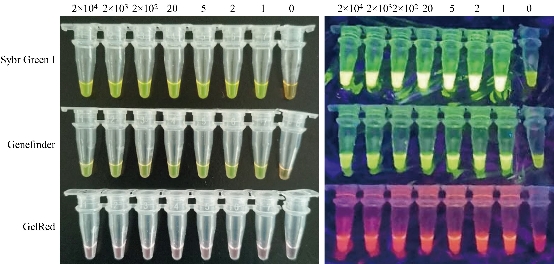
Fig. 1 LAMP assays on RNA reference materials with three chromogenic agents. Left: Observed with the naked eyes, bright green indicates a positive reaction for all the positive samples from 2×104 copies to 1 copy of RNA, and orange (or light green) a negative reaction for the negative control (0 copy) for Sybr Green I and GeneFinder staining; and violet for all the samples from 2×104 copies to 0 copy of RNA for GelRed staining. Right: Observed under the ultraviolet light, bright green indicates a positive reaction for all the RNA samples from 2×104 copies to 1 copy of RNA, and dark green a negative reaction for the negative control (0 copy) for Sybr Green I and GeneFinder staining; and bright violet indicates a positive reaction for all the positive samples from 2×104 copies to 1 copy of RNA, dark violet a negative reaction for the negative control (0 copy) for GelRed staining.
Seven positive samples with 2×104, 2×103, 2×102, 20, 5, 2 and 1 copies of RNA, respectively diluted from the RNA reference materials and one negative sample with 0 copy of RNA were tested using Sybr Green I, GeneFinder or GelRed as the chromogenic agents. The results indicated that all the positive samples from 2×104 copies to 1 copy of RNA were detected with the three chromogenic agents (Fig. 1). For the Sybr Green I staining, all the reaction products of the positive samples showed bright green observed with the naked eyes and under the ultraviolet light, while the negative control showed orange or light green observed with the naked eyes and dark green under the ultraviolet light. For GeneFinder staining, similar to Sybr Green I, all the reaction products of the positive samples showed bright green observed with the naked eyes and bright green under the ultraviolet light, while the negative control showed orange or light green observed with the naked eyes and dark green under the ultraviolet light. For GelRed staining, all the products of the samples showed violet and can’t be distinguished positive or negative with the naked eyes, while under the ultraviolet light, they can be distinguished with the products of the positive samples showing bright violet and the negative control showing dark violet. The results on RNA reference materials indicated that one copy of the RNA was detected with the optimized reaction, showing the high sensitivity of the SARS-CoV-2 detection kits.
LAMP assays on RNA samples from pseudo-virus
As shown in Figure 2, six positive RNA samples extracted from 104, 103, 102, 10, 2 and 1 copies of pseudo-virus, respectively were tested using the detection kits in which GeneFinder was used as the chromogenic agent. The results indicated that all the positive samples including the one extracted from 1 copy of pseudo-virus were detected. All the reaction products of the positive samples showed bright green observed with the naked eyes and under the ultraviolet light, while the reaction product of the negative sample showed orange or light green observed with the naked eyes and dark green under the ultraviolet light. The results on RNA samples from pseudo-virus indicated that one copy of the pseudo-virus was detected with the optimized reaction, showing the high sensitivity of the SARS-CoV-2 detection kits.
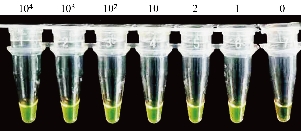
Fig. 2 LAMP assays on RNA samples from pseudo-virus using GeneFinder as the chromogenic agent. Bright green indicates a positive reaction for all the positive samples from 104 to 1 copy of pseudo-virus and orange (or light green) a negative reaction for the negative sample (0 copy).
LAMP assays on RNA samples from COVID-19 patients
As shown in Figure 3, four SARS-CoV-2 nucleic acid detection measurement audit samples prepared from the inactivated clinical samples were tested using the SARS-CoV-2 detection kits in which Sybr Green I was used as the chromogenic agent. The results indicated that three positive clinical samples including the low concentration sample No. 2016 and the positive control (RNA reference materials) were detected positive as the reaction products showing bright green observed with the naked eyes, while the negative clinical sample (No. 2013) and the negative control (DEPC-treated water) were tested negative as the reaction products showing dark green observed with the naked eyes.

Fig. 3 SARS-CoV-2 detection from COVID-19 patient samples using Sybr Green I as the chromogenic agent. Bright green indicates a positive reaction for all the positive samples including the clinical samples No. 2014, 2015, 2016 and the positive control (+), and orange (or light green) a negative reaction for the clinical sample No. 2013 and the negative control (-).
Conclusions
The SARS-CoV-2 nucleic acid detection kits based on colorimetric LAMP technology were provided for clinical diagnosis, which is of great significance to the surveillance and detection of COVID-19 pandemic. The efficacy of the kits based on the optimized reaction system was tested on RNA reference materials, pseudo-virus and clinical samples of SARS-CoV-2. As a result, as low as one copy of RNA molecules per reaction could be detected, showing the potential of higher sensitivity than the original reaction system containing primer group NA, which can detect the lowest RNA of 120 copies per reaction [17]. Compared with the commonly used molecular detection methods, the isothermal amplification method can be performed without complicated instruments and operations, so it is easy to be used under the insufficient conditions of complicated instruments and testing personnel. The efforts made here will have the potential to speed up the detection of suspected cases, reduce false-negative rate and avoid missed detection thus providing assistance for the diagnose and treatment of COVID-19. As we know, the sensitivity of nucleic acid detection is affected by many factors, including the disease course, specimen collection, detection process, detection reagent and other factors [6-10]. The results reported here were still preliminary and more experiments are needed on the detection limit, reproducibility, stability and specificity of the kits. In addition, higher sensitivity often means higher false-positive results, which will also bring the hidden problems and costs for the clinical diagnosis [18]. Therefore, the false-positive rate of the kits should also be avoided as other molecular detection operations.
Acknowledgements
This work was supported by Key Basic Research Program of China (No. 2017YFA0205304), Nature Scientific Foundation of China (No. 81602184), and Medical Engineering Cross Project of Shanghai Jiao Tong University (YG2017ZD12). This work was also supported by “the Belt and Road” young scientist exchange program of the Science and Technology Commission of Shanghai (Grant No. 18410741600).
Conflict of Interests
The authors declare that no competing interest exists.
References
Copyright© Xueling Li, Jing Tian, Yan Xu, Xiaomin Chen, Zexi Liu, Hui Liang, Caihong Jin, and Daxiang Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.