Research Article
A Rapid and High-Sensitive Real-Time Reverse Transcription-Polymerase Chain Reaction Assay Used for the Detection of Severe Acute Respiratory Syndrome Coronavirus 2
Xiaomin Chen 1, 2, Yan Xu 1, Jing Tian 1, Xueling Li 1, Hui Liang 1, Ang Gao 1, 2, Zexi Liu 1, Dicheng Yang 1, Qi Shen 1, Daxiang Cui 1, 2 *
1 National Engineering Research Center for Nanotechnology, Shanghai, China.
2 Institute of Nano Biomedicine and Engineering, Shanghai Engineering Research Centre for Intelligent Diagnosis and Treatment Instrument, Department of Instrument Science and Engineering, School of Electronic Information and Electrical Engineering, Shanghai Jiao Tong University, Shanghai, China.
* Corresponding authors. E-mail: dxcui@sjtu.edu.cn
Received: Aug. 24, 2020; Accepted: Oct. 27, 2020; Published: Oct. 27, 2020
Citation: Xiaomin Chen, Yan Xu, Jing Tian, Xueling Li, Hui Liang, Ang Gao, Zexi Liu, Dicheng Yang, Qi Shen, and Daxiang Cui, A Rapid and High-Sensitive Real-Time Reverse Transcription-Polymerase Chain Reaction Assay Used for the Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Nano Biomed. Eng., 2020, 12(4): 311-315.
DOI: 10.5101/nbe.v12i4.p311-315.
Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a public health emergency of international concern. Real-time reverse transcription-polymerase chain reaction (RT-PCR) is widely used as the gold standard method for the diagnosis of SARS-CoV-2 infection. However, the reliability of current real-time RT-PCR assays is questioned due to some false-negative reports. In this study, we improved the real-time RT-PCR method based on three target regions (ORF1ab, E, and N) of SARS-CoV-2. Results showed that real-time RT-PCR assays herein could complete detection within one hour after viral RNA preparation and had high sensitivity down to 5 copies of viral RNA. In addition, six clinical specimens were detected to evaluate the availability of this method. Among them, four samples were 3-plex SARS-CoV-2 positive and two were negative by real-time RT-PCR. The sensitivity was 100% (4/4), and specificity was 100% (2/2). These results demonstrate that we develop a rapid and high-sensitive real-time RT-PCR method for SARS-CoV-2 detection, which will be a powerful tool for COVID-19 identification and for monitoring suspected patients.
Keywords: COVID-19, SARS-CoV-2, RT-PCR, Diagnostic, Coronavirus
Introduction
The coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread around the world and has become a public health emergency of international concern [1, 2]. To date (20 August 2020), up to 22,213,869 COVID-19 cases have been confirmed, including more than 781,677 deaths [3]. In the absence of vaccines and antivirals, early diagnosis and quarantine are effective measures to cure patients and control the spread of COVID-19. The viral nucleic acid detection by real-time reverse transcription-polymerase chain reaction (RT-PCR) based on the TaqMan Probe assay is widely employed as the gold standard method for the diagnosis of SARS-CoV-2 infection [4, 5]. Advantages of real-time RT-PCR assays are high-throughput and sensitive, and even to quantify viral loads in patients based on a standard curve for monitoring disease progression [6]. Since the global outbreak of COVID-19, real-time RT-PCR assays based on various prime/probe sets are quickly developed for SARS-CoV-2 detection [7-10]. However, their reliability is questioned due to the presence of false-negative results in some patients and positive results in some confirmed cases after recovery [11-14]. Thus, it is very important to further optimize and improve current RT-PCR assays for better screening suspected patients and preventing COVID-19 spread. SARS-CoV-2 is an enveloped positive-sense single-stranded RNA (+ ssRNA) virus [1, 7]. Phylogenetic analyses reveal that SARS-CoV-2 belongs to β-coronavirus and its nucleotide sequence is close to bat-derived SARS-like coronaviruses [15, 16]. In order to distinguish from other high-pathogenic viruses, avoid viral mutation, and even detect the low viral load, SARS-CoV-2 is usually identified by multiple RT-PCR targeting different viral regions [4]. In this study, we optimized the real-time RT-PCR assay using four primer/probe sets targeting the open reading frame 1ab (ORF1ab), envelope protein (E), and nucleocapsid protein (N) regions of SARS-CoV-2 to improve COVID-19 detection. We also evaluated the sensitivity of this method. In addition, six clinical specimens were analyzed for the availability of this method.
Experimental
Preparation of the artificial SARS-CoV-2 gene
The synthesized sequence of the ORF1ab, E, and N gene of SARS-CoV-2 (BIOLIGO, Shanghai, China) were used to optimize real-time RT-PCR test. The SARS-CoV-2 RNA transcribed in vitro Reference Material (Shanghai Institute of Measurement and Testing Technology, Shanghai, China) containing ORF1ab (13321-15540, GenBank No.MT027064.1), the full-length E gene and N gene of SARS-CoV-2 quantitated by digital PCR were used to evaluate the sensitivity of real-time RT-PCR.
RNA extraction
Total RNA was extracted from the inactivated clinical specimens from Shanghai Center Clinical Laboratory (Shanghai, China) using the TaKaRa MiniBEST Viral RNA/DNA Extraction Kit Ver.5.0 (Takara, Japan) following manufacturer’s instructions. The extracted RNA was stored at -80 oC.
Primers and probes
Four high-specific primer/probe sets targeting the ORF1ab, E, and N regions of SARS-CoV-2 issued by China Center for Disease Control and Prevention (CDC) [7] and Corman et al. [8] were synthesized by BIOLIGO (Shanghai, China). Target 1 (ORF1ab), forward: 5’-CCCTGTGGGTTTTACACTTAA-3’, reverse: 5’-ACGATTGTGCATCAGCTGA-3’, probe: 5’-FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BQ1-3’; Target 2 (E), forward: 5’-ACAGGTACGTTAATAGTTAATAGCGT-3’, reverse: 5’-ATATTGCAGCAGTACGCACACA-3’, probe: 5’-FAM-ACACTAGCCATCCTTACTGCGCTTCG-BQ1-3’; Target 3 (N1), forward: 5’-GGGGAACTTCTCCTGCTAGAAT-3’, reverse: 5’-CAGACATTTTGCTCTCAAGCTG-3’, probe: 5’-FAM-TTGCTGCTGCTTGACAGATT-BQ1-3’; Target 4 (N2), forward: 5’-CACATTGGCACCCGCAATC-3’, reverse: 5’-GAGGAACGAGAAGAGGCTTG-3’, probe: 5’-FAM-ACTTCCTCAAGGAACAACATTGCCA-BQ1-3’.
Real-time RT-PCR assay
Real-time RT-PCR assay was developed using the One Step PrimerSciptTM RT-PCR kit (Takara, Japan). A 25 μL reaction mixture contained 12.5 μL of 2X PCR Buffer, 0.5 μL of reverse transcriptase, 0.5 μL of Taq DNA Polymerase, 200 nM of probes, 1 μL of primers, and 5 μL of RNA. Real-time RT-PCR was performed using Stratagene Mx3000P (Agilent, Palo Alto CA, USA). The optimal reaction procedure was 42 oC for 5 min, 95 oC for 10 s, followed by 40 cycles of 95 oC for 5 s and 60 oC for 30 s.
Statistical analysis
Data were presented as the mean + standard deviation (SD). The Student’s t-test was used in quantitative data analysis, and P < 0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed with SPSS 20.0 software (SPSS, Inc., Chicago IL, USA).
Results and Discussion
Optimizing the real-time RT-PCR assay
Many commercial nucleic acid detection kits based on real-time RT-PCR assays have been widely used for SARS-CoV-2 detection, but there are differences in their detection capability of weak positive samples [17, 18]. Currently, time-consuming and insensitive methods are not able to meet the requirement of large-scale molecular diagnosis of suspected patients and asymptomatic patients. Thus, to shorten the viral detection, we first performed real-time RT-PCR assays at three different thermal cycling time. As shown in Fig.1, the highest amplification of ORF1ab, E, and N2 at 400 nM of primers occurred in 35 s per thermal cycle, and N1 occurred in 25 s. Due to a multiple test, the optimal time per thermal cycle was determined at 35 s, meaning the whole real-time RT-PCR assay spent one hour completing detection. In order to increase the amplification products by real-time RT-PCR, we performed the reaction at different primer concentration from 200 nM to 400 nM. As presented in Fig. 2, the highest amplification of ORF1ab occurred at 400 nM of primers, and the other three genes had no significant differences in the range of 200 to 400 nM. Therefore, the optimal primer concentration of ORF1ab was 400 nM, and others adopted in were 200 nM.
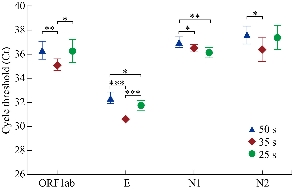
Fig. 1 Effect of thermal cycling time on real-time RT-PCR assays. Concentration of primers was 400 nM. *P < 0.05, **P < 0.01, ***P < 0.001.
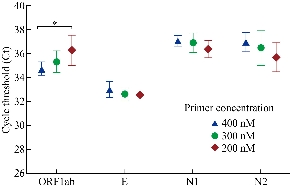
Fig. 2 Effect of primer concentration on real-time RT-PCR assays. *P < 0.05.
Sensitivity of the real-time RT-PCR assay
The performance of the aforementioned real-time RT-PCR assay was evaluated using ten-fold serial dilutions of the viral RNA ranging from 5x104 copies to 5 copies. As illustrated in Fig. 3, the cycle threshold (Ct) of real-time RT-PCR was linearly linked to the number of observed viral RNA, and regression analysis produced a determination coefficient (R2) of 0.941~0.992. The World Health Organization (WHO) posts various high-specific primer/probe sets for SARS-CoV-2 detection developed at different institutions [4]. However, assays among these primer/probe sets have different ability to detect SARS-CoV-2 [19, 20]. Chantal et al. [19] found that the most sensitive primer/probe sets in the same RT-PCR reagents and conditions are Corman E (Charité-Universitätsmedizin Berlin Institute of Virology), HKU-ORF1 (Hong Kong University), HKU-N (Hong Kong University), CCDC-N (China CDC), 2019-nCoV_N1 (United States CDC), and 2019-nCoV_N3 (US CDC), which could partially detect SARS-CoV-2 at 1 (25%) and 10 (25-50%) copies of viral RNA. Different from reaction conditions described by Chantal et al. [19], four primer/probe sets in this study also represented different amplification efficiency. The reportable range of ORF1ab was 5~5 x104 copies (Fig. 3A), and the limit of detection (LoD) of real-time RT-PCR using ORF1ab analyzed with twelve replicates was 5 copies of viral RNA (12/12) (Fig. 4A). The E assay also exhibited high sensitivity (Fig. 3B), but it was less sensitive than ORF1ab (Fig. 4B). In fact, LoD of real-time RT-PCR using E primer/probe set was more than 5 copies (5/12). Compared to the ORF1ab and E assays, the N assay was not reliable at low SARS-CoV-2 amounts. Among two N gene assays, the N1 primer/probe set was more sensitive than N2 set (Fig. 3C and D). To further confirm the improved real-time RT-PCR method, we utilized three primer/probe (ORF1ab, E and N1) sets in the subsequent studies.
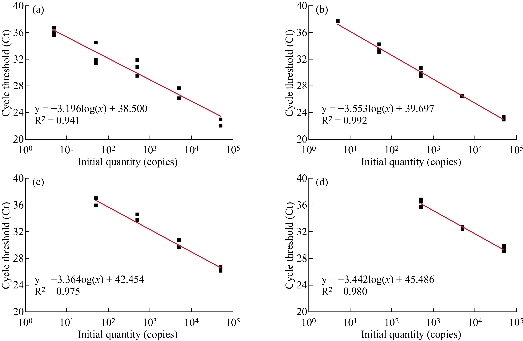
Fig. 3 Sensitivity analysis of real-time RT-PCR. The reportable range of real-time RT-PCR using (a) ORF1ab, (b) E, (b) N1, and (d) N2 primer/probe sets were determined. Data were representative of three independent experiments.
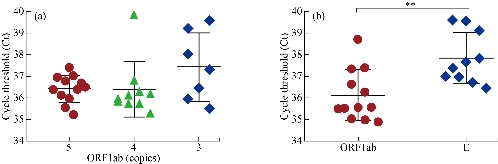
Fig. 4 (a) The limit of detection (LoD) of real-time RT-PCR based on ORF1ab primer/probe set. (b) Analysis of real-time RT-PCR using ORF1ab and E primer/probe sets to detect 9 copies of viral RNA. Data were analyzed with twelve replicates. **P < 0.01.
Clinical sample detection
Six clinical specimens were detected using the optimal real-time RT-PCR assay based on ORF1ab, E, and N1 primer/probe sets. Among them, four 3-plex positive samples were confirmed to be infected with SARS-CoV-2, while two were negative (Table 1). The sensitivity was 100% (4/4), and specificity was 100% (2/2). In addition, the results also showed that assays using ORF1ab or E primer/probe set performed better than those using N1 set (Table 1). Similar to most commercially available and laboratory-developed assays, we recommend that a 3-plex or 2-plex positive outcome is defined as SARS-CoV-2 positive, and not detected test result is negative. As for single-plex result, it is regarded as inconclusive and subsequently retested for the confirmation. Of course, a large scale of clinical study is necessary to prove the accuracy of this improved real-time RT-PCR method.
Table 1 clinical samples detected by real-time RT-PCR
|
Sample |
Ct Value |
Results |
||
|
ORF1ab |
E |
N1 |
||
|
1 |
- |
- |
- |
- |
|
2 |
30.20 |
31.71 |
35.60 |
+ |
|
3 |
- |
- |
- |
- |
|
4 |
28.87 |
29.94 |
33.21 |
+ |
|
5 |
28.16 |
28.79 |
32.87 |
+ |
|
6 |
35.31 |
33.95 |
38.84 |
+ |
-: Negative; +: Positive.
Conclusions
In this study, we have developed a rapid and high-sensitive real-time RT-PCR method based on the ORF1ab (China CDC), N1 (China CDC), and Corman E primer/probe sets to improve the current SARS-CoV-2 detection. This real-time RT-PCR assay can rapidly complete detection within one hour after viral RNA preparation. Moreover, the optimal multiplexed real-time RT-PCR assay exhibits high sensitivity down to 5 copies of viral RNA. In addition, our findings indicate that ORF1ab and E assays have a higher analytical sensitivity than N assays, which means that N assays are not a reliable confirmatory assay at low SARS-CoV-2 amounts.
Acknowledgements
This work was supported by Key Basic Research Program of China (No.2017YFA0205304), Nature Scientific Foundation of China (No.81602184), and Medical Engineering Cross Project of Shanghai Jiao Tong University (YG2017ZD12). This work was also supported by “the Belt and Road” young scientist exchange program of the Science and Technology Commission of Shanghai (Grant No. 18410741600).
Conflict of Interests
The authors declare that no completing interest exists.
References
Copyright© Xiaomin Chen, Yan Xu, Jing Tian, Xueling Li, Hui Liang, Ang Gao, Zexi Liu, Dicheng Yang, Qi Shen, and Daxiang Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.