Research Article
Molecular Study of Colibacillosis Susceptibility in Calves and Lambs
Hawraa Judi 1*, Rawaa Judi 2, Abdul-Kareem Saqban 1
1 Department of Microbiology, College of Veterinary Medicine, Al-Qasim Green University, Iraq.
2 Laboratory Division, Babylon Veterinary Teaching Hospital, Iraq.
* Corresponding author. E-mail: hawraakareem1991@gmail.com
Received: Feb. 14, 2020; Accepted: Apr. 13, 2020; Published: Apr. 27, 2020
Citation: Hawraa Judi, Rawaa Judi, and Abdul-Kareem Saqban, Molecular Study of Colibacillosis Susceptibility in Calves and Lambs. Nano Biomed. Eng., 2020, 12(2): 153-159.
DOI: 10.5101/nbe.v12i2.p153-159.
Abstract
Single nucleotide polymorphism (SNP) is hereditary change in a DNA sequence that occurs when a single nucleotide in a genome is modified; SNPs are usually considered to be point mutations that have been evolutionarily successful enough to recur in a significant proportion of the population of a species. The ability of certain individuals to respond appropriately to Toll-like receptor (TLR) ligands may be impaired by single-nucleotide polymorphisms (SNPs) within TLR genes, terminating in a modified susceptibility to infectious or inflammatory disease that might contribute to the pathogenesis of complex diseases. Out of the 400 clinical samples of blood collected from calves and lambs suffering from diarrhea, the samples originated from Babylon Veterinary Teaching Hospital and Veterinary Clinics during the period from October 2018 to January 2019, 200 samples were suffering from diarrhea (100 samples of calves and 100 samples of lambs), and 200 samples were the control groups. Genotyping of TLR4 polymorphisms was carried out by using amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) technique. For calves’ TLR4 (rs8193046) gene polymorphisms, the results showed there were significant differences in genotypes and alleles frequencies between the diarrheic cases and control groups (p < 0.05). GG homozygous genotype was overrepresented among diarrheic calves. The frequencies of G allele was higher in diarrheic calves compared to control groups. The calves’ GG homozygous genotype was significantly associated with increased susceptibility to Colibacillosis in calves. For lamb-TLR4 (rs160202325) gene polymorphism, AG heterozygous genotype was overrepresented among the cases as of 20 (52.63%), whereas AA was obviously more presented among control individuals as of 6 (60%). There were no difference between diarrheic cases and control groups for allele frequencies.
Keywords: Diarrhea, Gene polymorphism, TLR4, Calves, Lambs, ARMS-PCR
Introduction
Colibacillosis, an infection with Escherichia coli (E. coli) is a major cause of mortality among young calves [1, 2]. E. coli is a gram-negative bacterium belonging to the family of Enterobacteriaceae. This bacterium is motile and can produce multiple enzymes and proteins associated with its growth and development [3]. E. coli is normally present in the intestine of warm-blooded animals and humans. Although present in smaller amounts than the obligate anaerobes, this gram-negative bacillus is the predominant facultative anaerobe in the normal intestine [4]. Because of gain or loss of functions in genes, commensal E. coli may receive different types of virulence factors and thereby become pathogenic [5]. Route of infection might be endogenous or exogenous. Infections by E. coli are differentiated in 2 main types: Intestinal disease (e.g. diarrhea) and extra-intestinal disease (e.g. urinary tract infections, meningitis) [6]. Pathogenic E. coli is one of the etiologic agents of diarrhea and is known as diarrheagenic E. coli (DEC) which is of 7 types: Enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), Shiga toxin-producing E. coli (STEC, e.g. enterohemorrhagic E coli (EHEC)), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), diffusely adherent E. coli (DAEC), and adherent invasive E. coli (AIEC) [7]. Bacterial pathogens stimulate the inflammatory response in the gastrointestinal mucosa; although this can occur through different mechanisms, the final common outcome is the release of cytokines, chemokines and the recruitment of inflammatory cells [8]. Pro-inflammatory cytokines such as tumor necrosis factor- α (TNF-α), interleukin-1 (IL-1) and interleukin-6 (IL-6) have been noted to be the main ingredients that are necessary to initiate systemic inflammatory response [9]. Nucleotide-binding oligomerization domain (NOD) receptors, Toll-like receptors (TLRs) and pathogen-associated molecular patterns (PAMPs) have important roles in these complicated interactions. Extracellular and intracellular pattern recognition receptors (PRRs) recognize PAMPs, which stimulate inflammatory signaling cascades, for example, NF-κB and MAPKs pathways [10]. TLR signaling through the adaptor protein MyD88 can result in the activation of both MAPKs and NF-κB. NF-κB and MAPKs transduction cascades are evolutionarily ancient signaling pathways that are closely related to the defense against infection. The NF-κB complex consists of the IKKα and IKKβ catalytic subunits and the IKKγ regulatory subunit. Like NF-κB proteins, IKKα and IKKβ undergo homo- and heterodimerization [11]. TLR cytoplasmic domains resemble that of the IL-1 receptor, and as such they are called Toll-IL-1-receptor (TIR) domains. Following ligand-mediated dimerization, TLR recruits an adaptor protein MyD88, which then assembles a signalsome containing IRAK, TRAF6 and ECSIT [12]. The aim of the current study was to study the association between TLR4 single nucleotide polymorphisms (SNPs) with susceptibility or resistant to Colibacillosis in calves and lamb.
Experimental
Collection of specimens
During a time span from October 2018 to January 2019, out of 400 clinical specimen from blood were collected from diarrheic calves and lambs at Veterinary Teaching Hospital and Private Clinic.
Polymerase chain reaction
The ARMS-PCR technique was performed to detect TLR-4 gene polymorphism from lambs and calves’ blood sample. This method was carried out according to method described by Rosa et al. [13] (Table 1 and 2).
Table 1 ARMS-PCR primers for lambs’ TLR4 with their sequence and amplicon size with SNP
|
ARMSPCR primer |
Sequence |
Product size |
SNP |
|
Allele A primer Forward |
AGAGCACCTATGATGCCTTTGTA |
526 bp |
rs160202325 |
|
Allele G primer Forward |
AGCACCTATGATGCCTTTGTG |
524 bp |
|
|
Common reverse primers |
GGGGATTCTCCTCCTCAGGT |
||
Table 2 ARMS-PCR primers for calves’ TLR4 with their sequence and amplicon size with SNP
|
ARMS PCR primer |
Sequence |
Product size |
SNP |
|
Allele A Primer Forward |
GCATGCAGGAAGACACCG |
590 bp |
rs8193046 |
|
Allele G Primer Forward |
GCATGCAGGAAGACACCA |
590 bp |
|
|
Common Reverse Primers |
GCCCTCCTGAATAGCTAGGG |
||
DNA extraction
Genomic DNA from blood samples was extracted by using a DNA extraction kit (frozen blood) of Geneaid. USA, Then, extracted DNA was estimated by nanodrop device at 260/280 nm.
PCR master mix preparation
ARMS-PCR master mix conducted 2 reactions for each sample, including wild type allele master mix reaction and mutant type allele master mix reaction, using AccuPower PCR PreMix Kit according to company instructions (Table 3 and 4). The AS-PCR master mix reaction components mentioned above were placed in standard PCR tubes containing the PCR PreMix, with lyophilized materials containing all other components needed for PCR reaction such as Taq DNA polymerase, dNTPs, Tris-HCl pH 9.0, KCl, MgCl2, stabilizer, and tracking dye. Then, the tube placed in Exispin vortex centrifuged for 3 min, and then transferred in PCR thermocycler.
Table 3 ARMS-PCR master mix reaction for wild type allele
|
PCR master mix |
Volume (µL) |
|
Genomic DNA (5 - 50 ng) |
5 |
|
Generic primer (10 pmol) |
1 |
|
Wild type (T allele) primer (10 pmol) |
1 |
|
PCR water |
13 |
|
Total |
20 |
Table 4 ARMS-PCR master mix reaction for mutant type allele
|
PCR master mix |
Volume (µL) |
|
Genomic DNA (5 - 50 ng) |
5 |
|
Generic primer (10 pmol) |
1 |
|
Mutant (C allele) primer (10 pmol) |
1 |
|
PCR water |
13 |
|
Total |
20 |
PCR thermocycler conditions
PCR thermocycler conditions prepared by using conventional PCR thermocycler system were the same for each gene as shown in Table 5.
Table 5 PCR thermocycler conditions
|
ARMS-PCR step |
Temperature (℃) |
Time (sec) |
Repeat (cycle) |
|
Initial denaturation |
95 |
300 |
1 |
|
Denaturation |
95 |
30 |
30 |
|
Annealing |
60 |
45 |
|
|
Extension |
72 |
30 |
|
|
Final extension |
72 |
300 |
1 |
|
Hold |
4 |
Forever |
-- |
ARMS-PCR product analysis
ARMS-PCR products were analysed by loading in 1.5% agarose substance by using buffer, then stained with ethidium bromide, and observed under the UV transilluminator
Results and Discussion
Calves TLR4 gene polymorphism and Colibacillosis susceptibility
In this study, we investigated a possible association between TLR4 (rs8193046) polymorphisms and Colibacillosis in the local calve population of Babylon province, Iraq. The results of the present study demonstrated that the polymorphisms in calve TLR4 gene might affect the susceptibility to Colibacillosis and increased the risk of developing the disease. Fig. 1 shows the ARMS-PCR product analysis of calve TLR4 (rs8193046) polymorphism from blood samples of calves. The results of alleles frequencies of calve TLR4 (rs8193046) gene polymorphism in cases and control groups are presented in Table 7. The results for calve TLR4 (rs8193046) polymorphism showed that the G allele frequency was 48 (63.15%) in the infected calves and 6 (30%) in the control individuals, whereas A allele frequency was 28 (36.84%) in the study groups and 14 (70%) in control groups with a significant difference (p-value = 0.011) (Table 6). In our study, we found higher frequencies of G allele at calve TLR4 (rs8193046) in diarrheic calves compared to control groups. This allele was associated with increased susceptibility to Colibacillosis. The results of genotypes frequencies of calve TLR4 (rs8193046) gene polymorphism in diarrheic calves and control groups are presented in Table 7. Frequencies of the 3 genotypes (AA, GG and AG) of the cases and control groups are shown in Fig. 2. Results showed that GG homozygous genotype frequency was overrepresented among the cases, as of 17 (44.73%) which was higher than the other 2 genotypes AA 7 (18.42%) and AG 14 (36.84%) respectively. Therefore, GG homozygous genotype was the most frequent in patients. The calve TLR4 (rs8193046) GG homozygous genotype was significantly associated with increased susceptibility to Colibacillosis in calves. Calves with GG homozygous genotype were significantly overrepresented among diarrheic cases as of 17 (44.73%), compared with control groups as of 2 (20%), and had a 3.2381-fold increased risk of developing the disease than the other 2 genotypes (AG and AA), with p-value = 0.169, OR = 3.2381, and 95% CI = 0.3103 - 25.137 for GG genotype. In contrast, calves with AG heterozygous genotype were obviously better presented among the cases as of 14 (36.84%), compared with control groups as of 2 (20%), with p-value = 0.324, OR = 2.333, and 95% CI = 0.5814 - 11.5605 for AG genotype. On the other hand, calves with AA homozygous genotype were obviously better presented among the cases as of 7 (18.42%), compared with the control groups as of 6 (60%), with p-value = 0.013, OR = 0.1505, and 95% CI = 0.0333 - 0.6799 for AA genotype, as shown in Table 8. Our findings revealed that there were statistically significant differences between the study groups and the control groups for AA genotype frequencies regarding TLR4 (rs8193046) polymorphism (p-value < 0.05). However, there were no statistically significant differences between diarrheic calves and control groups for AG and GG genotype frequencies regarding TLR4 (rs8193046) polymorphism (p-value > 0.05). Moreover, there were statistically significant differences between the cases and the control groups for G and A allele frequencies regarding TLR4 (rs8193046) polymorphism (p-value < 0.05). Our findings were in agreement with the results reported by Rosa et al. [13] that genotype frequencies at rs8193046 between the study and the control groups showed statistically significant differences for AA genotypes. In our study, we found higher frequencies of G allele at TLR4 (rs8193046) in the cases. This allele was associated with increased susceptibility to Colibacillosis. The result was in agreement with that reported by Reeb et al. [14] who found the frequencies of G alleles at rs8193046 to be significantly higher in Colibacillosis than healthy groups, and the GG homozygous genotype being predominant for rs8193046 position. They also found that the frequency of GG genotype at TLR4 (rs8193046) polymorphic site was higher in the cases than in the control ones, suggesting an association with disease susceptibility; whereas the AA genotype was more common in control ones than in the cases, suggesting a protective association with Colibacillosis. Mishra et al. [15] and Seabury et al. [16] reported that genotype AA was associated with resistance to Colibacillosis, while genotype GG was associated with the risk of developing the disease.
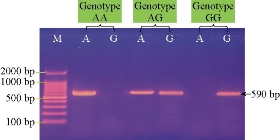
Fig. 1 Agarose gel electrophoresis image showing ARMS PCR product analysis of TLR4 Toll-like receptor 4 gene (Bos taurus) from blood samples of calves. Marker ladder: 2000 - 100 bp; Lane AG: Heterozygote genotypes; Lane AA: Homozygote wild type; Lane GG: Homozygote mutant type. The positive allele was shown at 590 bp ARMS PCR product size.
Table 6 Alleles frequencies of calve TLR4 (rs8193046) gene polymorphisms in infected and control groups
|
|
Case |
Control |
OR (95% CI)
|
P-value* |
||
|
Allele |
Count |
Proportion |
Count |
Proportion |
||
|
A |
28 |
36.84% |
14 |
70% |
0.2500 (0.0863 - 0.7244) |
0.011 |
|
G |
48 |
63.15% |
6 |
30% |
4 (1.3804 - 11.5907) |
|
Note: OR = Odd ratio; CI = Confidence intervals.
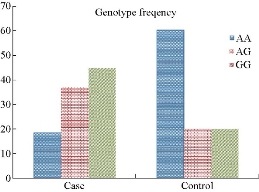
Fig. 2 Genotype frequencies of calve TLR4 gene polymorphisms in Colibacillosis cases and control groups.
Table 7 Genotype frequencies of calve TLR4 (rs8193046) gene polymorphisms in cases and control groups
|
Genotype |
Control No. 10 |
Case No. 38 |
OR (95% CI) |
P-value |
|
AA |
6 (60%) |
7 (18.42%) |
0.1505 (0.033 - 0.6799) |
0.013 |
|
AG |
2 (20%) |
14 (36.84%) |
2.3333 (0.5814 - 11.5605) |
0.324 |
|
GG |
2(20) |
17 (44.73%) |
3.2381 (0.3103 - 25.137) |
0.169 |
Lamb TLR4 gene polymorphism and susceptibility to Colibacillosis
Fig. 3 shows ARMS-PCR product analysis of lamb TLR4 (rs160202325) polymorphism from blood samples. Lamb-TLR4 (rs160202325) polymorphisms was genotyped using ARMS-PCR method as previously described by Mariotte et al. [17]. The results of allele frequencies of lamb-TLR4 (rs160202325) gene polymorphism in Colibacillosis and control groups are presented in Table 8 showing allele frequencies of the cases and control groups. In Table 9, the results for lamb TLR4 (rs160202325) gene polymorphism show that the A allele frequency was 38 (50%) in the infected animals and 15 (75%) in the control individuals, whereas G allele frequency was 35(50%) in the cases and 5 (25%) in the control individuals. The results of genotype frequencies of lamb TLR4 (rs160202325) gene polymorphism in cases, and control groups are presented in Table 9, and the frequencies of 3 genotypes (GG, AA and AG) of the cases and control groups are shown in Fig. 4. In Table 10, the results for lamb TLR4 (rs160202325) polymorphism show that AG heterozygous genotype frequency was overrepresented among the cases as of 20 (52.63%), which was higher than the other 2 genotypes as of 9 (23.68%) for GG and 9 (23.68%) for AA respectively, and had a 2.592-fold increased risk of developing Colibacillosis than the other 2 genotypes (GG and AA), with p-value = 0. 211, OR = 2.592, and 95% CI = 0.5814 - 11.5605 for AG genotype. The TLR4 (rs160202325) genotype was significantly associated with increased susceptibility to Colibacillosis infections. Lambs with AA homozygous genotype were obviously better presented among control individuals as of 6 (60%), compared with the cases as of 9 (23.68%), with p-value = 0.035, OR = 0.2069, and 95% CI = 0.0476 - 0.8996 for AA genotype. Similarly, individuals with GG homozygous genotype were obviously better presented among the cases as of 9 (23.68%), compared with the control as of 1 (10%), with p-value = 0.359, OR = 2.7931, and 95% CI = 0.3103 - 25.137 for AA genotype, as shown in Table 10. Our findings revealed there was no significant difference between the cases and the control groups for allele frequencies regarding lamb TLR4 (rs160202325) polymorphism. However, there were statistically significant differences in the frequency of TLR4 (rs160202325) genotype between cases with Colibacillosis and control groups (p-value < 0.05). Banos et al. found the frequencies of alleles at rs160202325 polymorphic site did not differ between Colibacillosis cases and control groups [18]. They also found AG heterozygous genotype was predominant for lamb TLR4 (rs160202325) and the frequency of AG genotype was significantly higher in the cases than in the control goups, suggesting an association with Colibacillosis susceptibility; whereas AA genotype was more common in the control ones than in the cases, suggesting a protective association with Colibacillosis. On the other hand, we found the frequency of AG genotype was higher in the cases as of 20 (52.63%) than in the controls as of 3 (30%), suggesting an association with Colibacillosis susceptibility; whereas the AA genotype was more common in healthy animals as of 6 (60%), suggesting a protective association with Colibacillosis. The results suggested that TLR4 gene polymorphism might play a role in Colibacillosis infection susceptibility and disease activity. TLR4 could respond to a variety of peptides derived from gram-positive organisms, a mutation in TLR4 could affect the immune response to various bacterial stimuli and potentially placed individuals at a significant risk of Colibacillosis [19]. Genetic variation in TLR4 has been reported to alter the susceptibility to various inflammatory and infectious diseases. Specifically, the well-known TLR4 (rs160202325) variant appeared to be associated with increased susceptibility to mastitis and Colibacillosis [20].
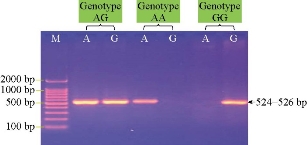
Fig. 3 Agarose gel electrophoresis image showing ARMS PCR product analysis of TLR4 Toll-like receptor 4 gene (Ovis aries) from blood samples of lambs. Marker ladder: 2000 - 100 bp; Lane AG: Heterozygote genotypes; Lane AA: Homozygote.
Table 8 Allele frequencies of lamb TLR2 (rs160202325) gene polymorphisms in cases and control groups
|
|
Control |
Case |
OR (95% CI) |
P-value* |
||
|
Allele |
Count |
Proportion |
Count |
Proportion |
||
|
A |
15 |
75% |
38 |
50% |
0.3333 (0.1101-1.0090) |
0.051 |
|
G |
5 |
25% |
35 |
50% |
3 (0.9911-9.0806) |
|
Note: OR = Odd ratio; CI = Confidence intervals.
Table 9 Genotype frequencies of lamb TLR4 (rs160202325) gene polymorphisms in diarrheic cases and control groups
|
Genotype |
Control No.10 |
Case No.38 |
OR (95% CI) |
P-value |
|
AA |
6 (60%) |
9 (23.68%) |
0.2069 (0.0476-0.8996) |
0.035 |
|
AG |
3 (30%) |
20 (52.63%) |
2.592 (0.5814-11.5605) |
0.211 |
|
GG |
1 (10%) |
9 (23.68%) |
2.7931 (0.3103-25.137) |
0.359 |
Table 10 Genotype frequencies of lamb TLR4 (rs160202325) gene polymorphisms in diarrheic cases and control groups
|
Genotype |
Control No.10 |
Case No.38 |
OR (95% CI) |
P-value |
|
AA |
6 (60%) |
9 (23.68%) |
0.2069 (0.0476-0.8996) |
0.035 |
|
AG |
3 (30%) |
20 (52.63%) |
2.592 (0.5814-11.5605) |
0.211 |
|
GG |
1 (10%) |
9 (23.68%) |
2.7931 (0.3103-25.137) |
0.359 |
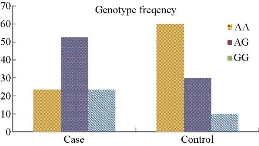
Fig. 4 Genotype frequencies of lamb TLR4 gene polymorphisms in Colibacillosis cases and control groups.
Conclusions
From this study, it was found that the TLR4 gene polymorphisms was clearly associated with susceptibility to Colibacellosis infection in calves and lambs.
References
Copyright© Hawraa Judi, Rawaa Judi, and Abdul-Kareem Saqban. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.