Research Article
Down-Regulation of fliL Gene Expression by Ag Nanoparticles and TiO2 Nanoparticles in Pragmatic Clinical Isolates of Proteus mirabilis and Proteus vulgaris from Urinary Tract Infection
Tahreer Hadi Saleh 1*, Saba Talib Hashim 1, Salma Nassrullah Malik 2, Bahaa Abdullah Laftaah AL-Rubaii 3
1 Department of Biology, College of Science, Mustansiriyah University, Baghdad, Iraq.
2 Department of Radiotherapy, Institute of Medical Technology, Baghdad, Iraq.
3 Department of Biology, College of Science, the University of Baghdad, Baghdad, Iraq.
* Corresponding author. E-mail: Tahreerhadi@gmail.com Tel.: +964 7709780888, +964 7506076334
Received: Aug. 18, 2019; Accepted: Oct. 14, 2019; Published: Oct. 16, 2019
Citation: Tahreer Hadi Saleh, Saba Talib Hashim, Salma Nassrullah Malik, and Bahaa Abdullah Laftaah AL-Rubaii, Down-Regulation of flil Gene Expression by Ag Nanoparticles and TiO2 Nanoparticles in Pragmatic Clinical Isolates of Proteus mirabilis and Proteus vulgaris from Urinary Tract Infection. Nano Biomed. Eng., 2019, 11(4): 321-332.
DOI: 10.5101/nbe.v11i4.p321-332.
Abstract
Ten isolates belonging to Proteus spp. were collected and obtained from Department of Biology, College of Science, the University of Baghdad. The diagnosis was done by polymerase chain reaction (PCR) technique using 16S rRNA gene and urease C gene. All isolates (100%( were sensitive to meropenem, imipenem, ciprofloxacin, gentamycin, amoxicillin, clavulanic acid and levofloxacin. These isolates also showed 60% sensitivity to cefixime and nitrofurantoin. However, both species of P. mirabilis and P. vulgaris showed the lowest sensitivity when treated with tetracycline (60%) and amikacin (20%). Cephalothin had a variable effect on the species under study as P. mirabilis isolates were 100% sensitive in comparison with the 80% sensitivity of P. vulgaris isolates. The antibacterial activities of Ag and TiO2 nanoparticles (NPs) were investigated. The minimum inhibitory concentration (MIC) value of Ag NPs against both species isolates was 10 mg/mL, while the MIC value of TiO2 NPs was 14 mg/mL against P. mirabilis and 15 mg/mL against P. vulgaris isolates. P .mirabilis isolates showed larger swarming diameter than P. vulgaris, but this motility phenomenon of P. vulgaris was arrested rapidly after incubation with sub-MIC of TiO2 NPs and Ag NPs comparatively with control. All isolates showed shifting to down-regulation in the fliL gene expression under the effect of the NPs using TiO2 NPs and Ag NPs. In conclusion, down-regulation of the fliL gene expression is directly linked to the inhibition of swarming movement of Proteus species. We encourage using these inhibitors (after tests to ensure minimal toxicity to human) in combination with antibiotics to ensure bactericidal /bacteriostatic effect to treat Proteus infections.
Keywords: P. mirabilis; P. vulgaris; Swarming phenomenon; fliL gene expression; TiO2 NPs; Ag NPs
Introduction
The genus of Proteus classification belongs to the family enterobacteriaceae and the swarming phenomenon is the most recognizable characteristics of Proteus spp compared to other members of the family [1]. This genus is gram negative pathogen, habiting in the intestinal tract of both animals and human, sewage, manure and environments [2]. Proteus vulgaris is an opportunistic bacterial pathogen in this genus coming after Proteus mirabilis and ranking as the third cause of UTI infection after E. coli and Klebsiella pneumoniae [3]. Proteus spp have many virulence factors helping in their adhesion, growth, colonization and invasion into infected tissues and thus progressing of the pathogenesis. These virulence factors include flagella, capsule, fimbriae, outer membrane proteins, lipopolysaccharides (LPS), biofilm formation, and several enzymes such as, haemolysin, metalloprotease, amino acid deaminase, urease [4] and chondroitinase [5, 6]. A lot of data about antibiotic resistance are obtainable about Proteus spp [7]. Proteus spp are resistant to different types of antibiotics such as polymyxin and tetracycline. Furthermore, multidrug-resistant strains were observed to be resistant towards antibiotics including aminoglycoside, streptothricin, fluoroquinolone, β-lactams, phenicol and trimethoprim-sulfamethoxazole [8]. In general, the 6~10 tiny peritrichous flagella on the surface of Proteus spp have been conceived as a catalyst for the colonization and transmission from the initial site to new location, and this movement is defined as swarming [9]. The swarming phenomenon is considered an important virulence factor differing from swimming; it is a multi-cellular operation that occurs on solid surface and needs the discrimination of vegetative cells into a special form of cell type termed as swarmer cells [10]. The swarming phenomenon is a flagellum-dependent movement style associated with increasing chance of infectious diseases caused by Proteus spp under suitable conditions, such as bacteraemia, wound infections, meningitis in infants, rheumatoid arthritis and others [11]. Colonization of Proteus spp on surfaces such as urinary tract is encouraged by differentiation of swarmer individual cell, which is established by hesitancy in flagellar spinning when first contacting with surface by the bacteria [12]. In general, the bacterial flagellum is built from: Firstly the basal body, followed by the hook, and then the helical filament, and the assembly of all is harmonious with a finely controlled regulatory rotation [13]. A mutation, disorder or other events lead to variations in fliL gene, encoding a flagellum structural protein may lead to improper creation of swarmer cells, known as pseudo-swarmer cells, indicating the sharing of fliL protein in the surface sensing process under non-motivational conditions [12]. Nanotechnology is one of the sciences that deal with the production of nanoparticles which are very small in size, ranging from 1 to 100 nm and possessing a high surface area compared with huge examples. Nanoparticles can be synthesized from many metals including silver, platinum, titanium, gold, copper, etc. Nanoparticles have unique chemical, physical and biological properties [14]. Biogenic nanoparticles have been applied in bactericidal applications, which may be attributed to their biocompatibility and long-term stability [15]. Metal nanoparticles have become one of the hopeful alternatives to defeat the microbial resistance of MDR bacteria [16]. Mechanisms of these nanoparticles include metal ion release, and oxidative and non-oxidative stress being active at the same time [17]. These mechanisms caused membrane degradation, deterioration of cellular and so on [18]. Silver nanoparticles (Ag NPs) are among these inorganic agents, which is non-toxic, prepared from silver metal, and having bactericidal, antiviral and anti-fungal effects at low concentrations [19]. Titanium dioxide nanoparticles (TiO2 NPs) are another one with stability in their physical and chemical structure, and optical, electrical and biocompatible properties. They are considered safe and non-toxic as confirmed by the American Food and Drug Administration (FDA). They are used in many applications such as cosmetics, tooth pastes and detergents, showing powerful germicidal feature and removing unpleasant odours [20]. These nanoparticles have wide spectra of antimicrobial agents; therefore, they can be used as alternative treatments for many diseases caused by bacteria [21]. Titanium is usually used in treating some skin infections as a sun blocker among patients who suffer from dermal damage, due to its safety and better ability to absorb UV radiation than other nanoparticles [2]. It has been applied in environmental usage to remove contaminants from both air and water. It has also been used as a semiconductor photo-catalyst [23].
Experimental
Isolation, culture conditions, and identification
In this study, 10 isolates of pathogenic bacteria belonging to Proteus spp were obtained in slant tubes containing brain heart infusion agar from post graduate students (Department of Biology, College of Science, Baghdad University). These isolates were activated by re-culturing on different media for primary diagnosis performed according to morphological and biochemical tests and then maintained in brain heart infusion agar. The diagnosis was re-confirmed by PCR technique using specific set of primers’ composition for amplification of 16S rRNA gene, f5-'CACGCAGGCGGTCAATTAAG-3′ and r5-' TCTTTTGCAACCCACTCCCAT -3′ [24]. In order to amplify the specific urease C gene, another set of primers were employed to confirm that isolates of Proteus spp belong to P. vulgaris including f5-'CGCTTTGCGATGGCAAGTACAAGTAAC-3′ and r 5-'GCAAATTGAGTGACTTTGGCTGGACC-3′ [6]. In PCR tube, reaction was performed on a total volume of 25 μL mixture which contained 12 μL of Green PCR Master Mix, 1 μL of each primer, and 2 μL of DNA template. The rest volume was achieved with sterile deionized distilled water, then mixed well by vortex. The suitable PCR process for 16S rRNA gene amplification was initial denaturation, denaturation, annealing, and extension, at 95 oC for 5 min, 30 cycles at 95 oC for 30 sec, 58 oC for 30 sec, and 72 oC for 1 min respectively, where the final step (extension) was conducted at 72 oC for 10 min and then held at 4 oC. The above PCR process was identical to urease C gene amplification except that annealing temperature was 62 oC.
Anti-program susceptibility test
Antibiotic sensitivity
According to Kirby-Bauer disc diffusion manure, the antiprogram susceptibility profile was done on Mueller-Hinton agar [25]. Briefly, the isolates were left to be grown in BHI broth for 18 h at 37 ºC. All isolates were prepared to a turbidity referred to as 0.5 McFarland standard (108 cfu/mL) [26]. Fresh 100 μL of bacterial suspension was spreading well on Mueller-Hinton agar plate, and then antibiotics disc was applied on surface of each of the 5 plates and incubated for 24 h at 37 ºC. The diameters of clear halo zones of each individual disk were metric with millimetres, and sorted as resistant or sensitive grade according to the clarification list of CLSI [27] and based on comparison with antibiotic susceptibility analysis of E. coli ATCC 25922. All the antibiotics discs were offered by Oxoid, including levifloxacin 5 μg, cefixime 5 μg, ciprofloxacin 5 μg, tetracycline 30 μg, cephalothin 30 μg, meropenem 10 μg, imipenem 10 μg, gentamycin 10 μg, amikacin 30 μg, nitrofurantoin 30 μg, and amoxicillin + clavulanic acid 30 μg.
Minimum inhibitory concentration (MIC) determination of nanoparticles
0.5 gram of each of Ag NPs and TiO2 NPs (Sigma) were found to be of 100-nm particle size by transmission electron microscopy (TEM), dissolved in separated tubes containing 9 mL of distilled water (DW) and 50 µL of dimethyl sulfoxide (DMSO) which were added to facilitate the dissolving process. The volume was completed up to 10 mL by DW to get a concentration of 50 mg/mL as stock solution. Following that, from the stock solution different gradient concentrations were prepared. The antibacterial activity was investigated by agar well diffusion method using Mueller-Hinton agar plate. The wells were inoculated with microliters of nanoparticle samples which were prepared with concentrations ranging from 1 to 20 mg/mL separately. Then, the inhibition halo around wells in agar were monitored and metric in millimetres after plates were incubated at standard condition [28].
Investigation effect of nanoparticles as anti-swarming
Phenotypic effect
In Eppendorf tubes, 50 μL from the overnight culture of Proteus spp was mixed well with the sub-MIC watery solutions of Ag NPs and TiO2 NPs, separately, and then incubated at 37 °C for 24 h under monitoring. In the next incubation, 5 μL of the above culture was implanted on the centre of blood agar plates and allowed to incubate in the same condition as sated above. It was observed that the waves of swarming diameter started from the central spot of inoculation, were metric in millimetre, and matched with the control (bacterial plat swarming without nanoparticles’ treatment). The nanoparticles that made a movement of diameter wider than the control was categorized as stimulatory action; however, if the zone was smaller than the reference colony, it was labelled as inhibitory [29]
Genotypic effect
fliL gene expression analysis was monitored before and after treatment of Proteus spp isolates with Ag NPs and TiO2 NPs.
Incubation of bacteria with nanoparticles
The isolates were left to grow in BHI broth for 18 h at 37 ºC. Then, 100 μL (0.5 McFarland) of fresh bacterial suspension was transferred to the Eppendorf tubes containing sub-MIC of both nanoparticles independently, mixed well by pipetting for few 2-5 min and then incubated for 24 h at 37 ºC until used in the next step.
RNA extraction
The bacterial growth was collected and harvested from BHI, and then the total RNA of isolates was taken away according to instructions of TRIzol™ kit (Thermo Scientific, USA). All extraction was treated with the DNase for 1 h. The purification and concentration values were detected by using a NanoDrop™ 1000 spectrophotometer and a Quantus™ fluorometer (Promega, USA). In Eppendorf tubes, 1 μL of each RNA sample was mixed together with 199 μL of water-quality flour colorant and incubated in dark place at room temperature for 5-10 min.
cDNA synthesis, Evaluation of RT-PCR
The fliL gene expression shot was estimated by one step quantitative RT-qPCR using sets of primer: fliLF5'-GGTGATCGCCATTATTGCAG -3', fliLR5'-AGCGTAACGTGATCCCTATG-3'. And rpoA acting as housekeeping gene was F5'-GCGTGTTATAGCCCAGTTGA-3' and R5'-AGGCTGACGAACATCACGTA-3' [12]. The thermocycling requirements were 25 °C for 10 min, 37 °C for 2 h, and 85 °C for 5 min, employed to convert about 1 μg of total RNA into cDNA by cDNA reverse transcription kit containing RNase inactivate agent. For RT-PCRs, 50 μL for each reaction solution contained 2 μL (200 μM) of deoxynucleoside triphosphates (dNTPs), 1 μL (200 nM) of each primer, 1 μL of ThermoPol buffer and 2 μL (25 ng) of cDNA as template were mixed well with 1μL of Taq polymerase (5 IU). The experimental conditions were 95 °C for 3 min, 30 cycles at 95 °C for 1 min, 60 °C for 30 sec, and 72 °C for 30 sec, and 72 °C for 2 min; qRT-PCR was run with the SYBR green kit. For each reaction, the mixture was brought up to 25 μL as final volume with nuclease-free water, incorporating 2 × SYBR green master mix, 1 μL (200 nM) of each primer, and then pipetted well with 2 μL (25 ng) of cDNA as template. And to follow, the reaction was carried out on a Mic qPCR Cycler (Bio MolecularSystem, Australia) using GoTag qPCR Master MixGoTaq® 1-Step RT-qPCR System, (Promega, USA), following the optimized conditions: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 20 sec, and 60 °C for 60 sec. A separation curve interpretation was achieved for test to maintain harvest (95 °C for 20 sec, 60 °C for 60 sec, and 95 °C for 20 sec). The quantum of fold (change) was calculated using the 2-ΔΔCTformula: Folding = 2-ΔΔCT, ΔΔCT = ΔCT treated - ΔCT control, ΔCT = CT gene - CT housekeeping gene [30]. The threshold cycle (CT) method was employed in relative expressions calculation for quantitative RT-qPCR.
Statistical analysis
Data analysis in this article was completed by SPSS 16.0 (SPSS Inc., Chicago, IL, USA) and calculated by student’s t-test (t) and ANOVA test. P-value < 0.05 was deemed statistically significant.
Results and Discussion
Isolation, culture conditions and identification
The ten isolates of Proteus spp., obtained from post graduate students, were re-activated on BHI agar. Results of the primary diagnosis were that all the isolates (100%) were non-lactose fermenter, pale colonies on MacConky agar. Also, all the isolates exhibited the swarming phenomenon clearly when cultured on blood agar plates, which is the main characteristics to distinguish Proteus spp. from other members of Enterobacteriaceae. Our results using molecular techniques showed that all isolates in this study were Proteus species. Amplification of 16S rRNA gene and urease C gene were separated on agarose gel by electrophoresis with amplicon size 857 bp and 263 bp, respectively under UV-spector, confirming identification of the isolates as P. mirabilis and P. vulgaris, respectively (Fig. 1 and 2). Our findings are similar to those obtained by Al-Imam, et al. and Al-Saadi, et al. who used the same genes to identify P. mirabilis and P. vulgaris [6, 31]. On the other hand, Mukhtar et al. and Adnan et al. applied 16S rRNA PCR gene amplification technique for the identification of P. mirabilis [32, 33]. The latter technique has many advantages over phenotypic and biochemical techniques. It is extremely preserved within types and among species of the same genus, present in all bacteria and considered a fixed structural gene (little variation frequency) [34]. The lack of functional mutations in this gene leads to fine identification compared with other genes that may be exposed to different mutations which may cause dysfunction. And it is highly accurate compared with phenotypic assays by bacteriological and biochemical methods [35] which may be changed due to environmental conditions, growth conditions, temperature and pH levels, etc. [36]. Using PCR technique in bacterial diagnosis has been proved by all means to be a reliable, easy, rapid and accurate procedure for identification and diagnosis [37], overcoming the difficulties and unpleasant results compared with conventional methods in diagnosis. Since the conventional identification of bacteria is usually achieved by a series of biochemical tests, the major disadvantage of this approach is it takes long time and is expensive for laboratory. Its close relation with other individuals of Proteus genus and other enterobacteriaceae has also made the identification of Proteus spp. difficult.

Fig. 1 Gel electrophoresis of amplified 16S rRNA (857 bp) for Proteus spp. isolates on agarose (1%), TBE buffer (1×), stained with ethidium bromide. M: DNA ladder (100 bp); Lanes 1-10 were positive.
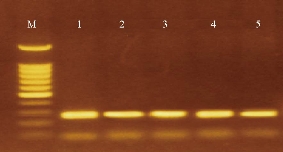
Fig. 2 Gel electrophoresis of amplified urease C (263 bp) for P. vulgaris isolates on agarose (1%), stained with ethidium bromide. M: DNA ladder (100 bp); Lanes 1-5 were positive.
Antibiotics susceptibility
The results are illustrated in Fig. 3, showing variable patterns of susceptibility against different antibiotics. The antiprogram pattern displayed that all isolates of P. mirabilis and P. vulgaris were sensitive (100%) to meropenem (MEM), imipenem (IPM), ciprofloxacin (CIP), gentamycin (GEN), amoxicillin + clavulanic (AUG) acid and levofloxacin (LEV). In addition, these isolates were 60% sensitive under the effect of cefixime (CEF) and nitrofurantoin (NIT). However, both species exhibited the lowest sensitivity patterns of 60% and 20% toward tetracycline (TET) and amikacin (AMK), respectively. On the other hand, cephalothin (CEP) had a variable effect on the two species as P. mirabilis isolates were 100% sensitive in comparison with 20% by P. vulgaris. It is interesting to find that our results were similar to other studies reported by several authors who noticed all isolates of P. vulgaris were sensitive against meropenem, ciprofloxacin, levofloxacin, gentamycin, cephalothin and amoxicillin + clavulanic acid. The swarmer cells showed an increase in their resistance to different antibiotics, but re-culturing these cells in broth media may cause them to be normal planktonic cells and to thereby exhibit different antibiotic susceptibility patterns [40]. The results of our study did not match with other reports which showed that vulgaris was 100% resistant to cephalothin and 93.3% to nitrofurantoin, but showed that 96.6%, 80%, and 65.7% of isolate were sensitive to ciprofloxacin, gentamycin, and amikacin, respectively [41]. Other studies of P. vulgaris isolates showed it was resistant only to ampicillin and cefuroxime [42]. However, extended spectrum beta-lactamases (ESBLs) play a role in Proteus spp. resistance to cefotaxime, ceftazidime, and ceftriaxone, as well as aztreonam, monobactam and the cephamycins (cefmetazole, cefoxitin and cefotetan), and the carbapenems (meropenem and imipenem) which are generally not hydrolysed by ESBLs [43]. Proteus spp. can win resistance to antibiotics such as ampicillin through plasmid and chromosomal beta-lactamase expression [44]. The shifting in the regulatory (responsible) genes of the beta-lactamase was observed to produce high activity of the enzyme and cause resistance to cefotaxime, ceftriaxone, cefuroxime and penicillin [45]. And the change might occur in penicillin binding proteins which are responsible for synthesis of cellular wall peptidoglycan [46]. There are many reasons for bacterial resistance, including modified enzyme production which is able to inhibit activity of antibiotics. The occurrence of safe mutation leads to change in the target site of antibacterial drugs, reducing permeability behaviour. Efflux pumps, R-plasmids, integrons and transposons are probably acquired from other microorganisms that are found in the same environment [47, 48]. Proteus spp. can be naturally resistant to oxacillin, benzylpenicillin and macrolides [49]. The antibiotic susceptibility tests illustrated that Proteus spp. had a variety style, which may be attributed to lipid lipoproteins, bilayer, polysaccharides and LPS found in the extra outer cytoplasmic membrane. Additionally, misuse and abuse of different antimicrobial agents in medicine (medical and veterinary) may lead to the spreading and development of antibiotic resistance genes strategies [50, 51]. Given all the information above, we can consider Proteus spp. as an ESBL producing bacteria due to their ability to produce a chromosomally encoded beta-lactamase. This conclusion is in agreement with results achieved by Bush et al. [52].
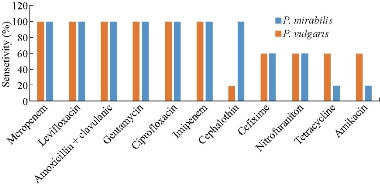
Fig. 3 Antiprogram profile of P. mirabilis and P. vulgaris isolates toward routinely antibiotic disks
Determination of antibacterial activity and MIC
The augmentation in bacterial resistance to antibiotics became an overall health question. A short time ago, nanoparticles have become an agent at odds with multidrug-resistant (MDR) bacteria. Many in-vitro and in-vivo results reported that metal nanoparticles had bactericidal activities towards a wide spectrum of bacterial species. However, in this study, the antibacterial activity for both Ag NPs and TiO2 NPs was investigated by dilution method in agar wells. The MIC values of Ag NPs against P. mirabilis and P. vulgaris isolates were detected at 10 mg/mL (Table 2 and 3). However, the highest effective concentrations of TiO2 NPs causing the total perishing of P. mirabilis was 14 mg/mL and 14-15 mg/mL for P. vulgaris isolates (Table 4 and 5). We can conclude that Proteus spp. isolates may be able to switch on some mechanical resistance associated with gradual increase in the concentration of TiO2 NPs less than in the case of silver, which caused the loss of all isolates. Our results were in disagreement with results about MIC values of Ag NPs against Proteus spp. at 10 μg/mL and 16 μg/mL as reported by Parveen et al. and Raheem et al. [53, 54], and at 0.015 mg/mL as investigated by Junevičius and co-workers [55]. But the MIC values reached to 50 mg/mL when Proteus spp. were treated with Ag NPs [56]. There has been no article available that detected the MIC values of titanium dioxide nanoparticles against Proteus spp. However, Chatterjee and co-workers observed that Proteus mirabilis treated with titanium dioxide nanoparticles gave the inhibition zone about 3.4 mm around colonies [57]. On the other hand, MIC values of titanium nanoparticles were 128 μg/mL [58] and 20 mg/mL, 72 mg/mL, and 100 mg/mL for Shigella dysenteries, multidrug-resistant E. coli, and Aeromonas hydrophilic, respectively [59]. The mechanism of action for both antibiotics and nanoparticles was the same in way of interference in the synthesis of macromolecules such as DNA, RNA, protein, as well as membrane destroyed [60]. However, the antibacterial mechanism of Ag NPs was described by many articles, including (i) Ag NPs can attach with cell membrane of bacteria causing disruption of its permeability and causing the figuration of many pores and slots in the cell membrane and then the influx of intra-cellular contents [61, 62]; (ii) free radicals and reactive oxygen species (ROS) such as H2O2 are liberated by Ag NPs on the superficies of nanoparticles and cause deactivate of DNA molecule by effectiveness in replication enzymes’ activity [63]; (iii) the Ag ions which are released by Ag NPs react with the thiol group of some bacterial proteins that are responsible for many important cellular functions and inactivation of them, such as damaged DNA molecules [64, 65]; and (vi) disruption of the ATP production [66]. However, the action of titanium nanoparticles against bacteria is dissolving the outer membranes of microbes due to the presence of hydroxyl groups that may cause change in the permeability of the cellular membrane and finely lead to the death of pathogen, which may be attributed to the ability of interaction with O2 and -OH are adsorbed on the surface to obtain OH and O2 free radical [67, 68]. The photocatalytic operation of titanium nanoparticles triggers the reduced expression of a wide range of genes encoded proteins set for regulatory control, signalling and progressive roles in tantamount with the next chosen events on cell wall texture, co-enzyme independent respiration, and ion homeostasis [69]. Also, titanium dioxide nanoparticles are able to cause damage of DNA, formation of reactive oxygen species (ROS) and superoxide radicals [70, 71]. Some reports demonstrated that exposing bactria to TiO2 photocatalysis quickly disrupted the regulatory signalling level, reduced the coenzyme-independent respiratory chains, downregulated the facility to take up and carry phosphorous and iron, and inhibited the ability of bio-synthesis and declination of heme Fe-S cluster [16, 72]. For example, TiO2 NPs display antimicrobial activity by different strategies given that the chance of expansion of reluctance against these nanoparticles is depressed [73]. However, in order to enhance the resistance against these metal nanoparticles, the bacteria may need to gain numerous gene mutations, which is not very likely [74]. On the other hand, Proteus spp. genome carries antibiotic resistance loci towards different mechanisms of antimicrobial agents and metals, including swarming mobility, biofilm formation, enzymatic detoxification and efflux systems by employing the PATRIC and PGAAP gene explanation services [75-77].
Table 1 MIC values for Ag NPs vs. P. mirabilis
|
Isolate |
Ag NPs concentration (mg/mL) |
|||||||||||||||||||
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 |
20 |
|
|
P. mirabilis 1 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. mirabilis 2 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. mirabilis 3 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. mirabilis 4 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. mirabilis 5 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
* Positive (+) means inhibition of bacterial growth.
* Negative (-) means bacterial growth.
Table 2 MIC values for TiO2 vs. P. mirabilis
|
Isolate |
TiO2 concentration (mg/mL) |
|||||||||||||||||||
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 |
20 |
|
|
P. mirabilis 1 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. mirabilis 2 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. mirabilis 3 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. mirabilis 4 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. mirabilis 5 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
* Positive (+) means inhibition of bacterial growth.
* Negative (-) means bacterial growth.
Table 3 MIC values for Ag NPs vs. P. vulgaris
|
Isolate |
Ag NPs concentration (mg/mL) |
|||||||||||||||||||
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 |
20 |
|
|
P. vulgaris 1 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. vulgaris 2 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. vulgaris 3 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. vulgaris 4 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. vulgaris 5 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
* Positive (+) means inhibition of bacterial growth.
* Negative (-) means bacterial growth.
Table 4 MIC values for TiO2 vs. P. vulgaris
|
Isolate |
TiO2 NPs concentration (mg/mL) |
|||||||||||||||||||
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 |
20 |
|
|
P. vulgaris 1 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. vulgaris 2 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. vulgaris 3 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. vulgaris 4 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
P. vulgaris 5 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
* Positive (+) means inhibition of bacterial growth.
* Negative (-) means bacterial growth.
The impact of nanoparticles on swarming phenomenon
Results showed all isolates varied in their behaviour of swarming. It was observed that P. mirabilis isolates’ motility ranged from 21 - 42 mm, larger and faster than P. vulgaris which ranged from 14 - 35 mm (p-value < 0.05). The restraint and obstruction role for sub-MIC of TiO2 NPs on the diameter of swarming waves’ motion of P. vulgaris isolates was observed as more active than the sub-MIC of Ag NPs of P. mirabilison (p-value < 0.05) (Table 5 and 6). These two sub-MIC might increase the impairment of flagellar synthesis or rotation and thus the activity in cellular movement of Proteus spp. It was concluded that the swarming was of dose-dependent pattern reduced by both TiO2 NPs and Ag NPs. There are many reasons behind the deviation in swarming patterns of Proteus spp. that may be associated with the bacteria themselves, such as strain variation, growth condition, source, incubation condition (the containing medium, pH, temperature, moisture, etc.), and expression of particular related swarming genes. Our results may be confirmed by Senior’s view that infectious diseases were encouraged more by P. mirabilis than by P.vulgaris due to their efficiency to produce many virulence factors such as protease and haemolysin, etc. that synchronized with the swarming motility and then moved out of urinary tract to another site in host tissues [78]. Most previous articles lacked the effect of TiO2 NPs and Ag NPs on the swarming phenomenon presented by Proteus spp., but there are some other articles dealing with the arresting effect of synthesized Ag NPs on rhl regulation system which controls the swarming movement exhibited by P. aeruginosa [79]. The synthesized Ag NPs demonstrated a reducing effect on the swarming behaviour of E. coli, P. aeruginosa PAO1, and K. pneumoniae [80]. Also, Ag NPs showed to have reduced about 98% activity of the swarming motility associated with the arresting effect in the expression of fliL flagellar gene in E. coli [81]. The swarming movement was the reason of combination sensory transduction and universal control operations. The swarming cells called for sensing and coupling of an assortment of cell-to-cell, environmental and intracellular signals, and involved adjusting expression of gene networks key to physiological and morphological alterations [82]. Alternations in gene flaA encoding flagellin protein [83], and flhDC gene contributed to the upregulation of flagellin protein production, C and wad genes needed for the core region of LPS [84]. Other genes implicated in flagellum construction drove the repression of Proteus rods’ segregation and inhibition of swarming.
Table 5 Effect of Ag NPs and TiO2 NP at sub-MIC on the swarming of P. mirabilis.
|
Isolates |
Swarming control (mm) |
Ag NPs sub-MIC (8 mg/mL) |
TiO2 NPs sub-MIC (10 mg/mL) |
|
P. mirabilis 1 |
21 |
13 |
12 |
|
P. mirabilis 2 |
27 |
22 |
19 |
|
P. mirabilis 3 |
40 |
32 |
30 |
|
P. mirabilis 4 |
44 |
35 |
32 |
|
P. mirabilis 5 |
42 |
23 |
20 |
Table 6 Effect of Ag NPs and TiO2 NP at sub-MIC on the swarming of P. vulgaris.
|
Isolates |
Swarming control (mm) |
Ag NPs sub-MIC (8 mg/mL) |
TiO2 NPs sub-MIC (10 mg/mL) |
|
P. vulgaris 1 |
14 |
9 |
8 |
|
P. vulgaris 2 |
18 |
13 |
10 |
|
P. vulgaris 3 |
26 |
23 |
21 |
|
P. vulgaris 4 |
31 |
28 |
23 |
|
P. vulgaris 5 |
35 |
30 |
24 |
Effect of nanoparticles on fliL gene expression
The results proved that incubation of isolates with both sub-MIC of TiO2 NPs and Ag NPs caused alternation in the expression manner of the fliL gene after 24 h. In this study, the Ct values of fliL gene in all isolates of P. mirabilis and P .vulgaris increased significantly (p-values < 0.05) after exposed to nanoparticles (Fig. 1 and 2), and the fold change 2-˄∆∆ ct was less than 1 (Table 7 and 8). The threshold cycle Ct method was employed in relative expressions calculation for quantitative RT-qPCR; all the results were compared to the control (which was not treated with any nanoparticles under the same experimental conditions). Hence, we could conclude that gene expression was 100% down-regulated, and our results may reveal a regulatory link between fliL gene and swarming prominence. In other words, the isolates may not have an antioxidant preservation strategies to remain off ROS created by TiO2 NPs and Ag NPs, which may be integrated with fliL gene and then reduce their regulation, that is to say, inhibition of movement is synchronized with the reduction of fliL gene expression. The down regulation may occur due to interfering TiO2 NPs and Ag NPs with different extra and entry cellular proteins such as flagellar protein and cause arrest in their movement function. As can be said, reasons beneath the lowering in gene expression may belong to the same antimicrobial mechanisms of nano-material against pathogens, including damage of bacterial membrane and cell wall, deterioration to bacterial proteins and internal parts of bacteria, ions’ liberation, and oxidative stress and DNA damage [85], or that ribosomal loss subunits ability to expression of cellular proteins [86]. Any disorder or any treatment with exogenous compounds, such as chemicals, may lead to change in the function of fliL gene, which is responsible for encoding a flagellar structural protein and causes creation of pseudo swarmer cells, suggesting the sharing of fliL in the surface sensing passage under no stimulating conditions [12]. FliL protein is necessary for swarming movement, but not for swimming motion [87]. The relative expressions calculation of quantitative RT-qPCR method for bacterial genes function was employed widely in different reports. Shehab and co-workers marked the alternation in stable-state mRNA levels of a gene towards numerous samples and described it as comparative to levels of a domestic control RNA [88].
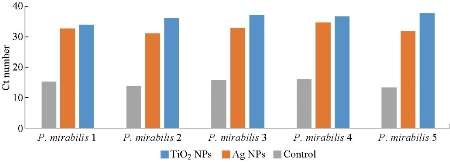
Fig. 4 Down-regulation in fliL gene expression of P. mirabilis exposed to sub-MIC of nanoparticles.
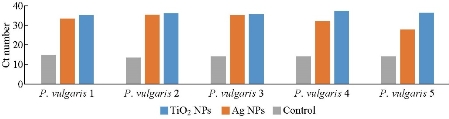
Fig. 5 Down-regulation in fliL gene expression of P. vulgaris exposed to sub-MIC of nanoparticles.
Table 7 The folding change of treated fliL gene expression in P. mirabilis
|
Isolates |
Folding change 2˄∆∆ct |
|
|
|
Sliver (8 mg/mL) |
Titanium (10 mg/mL) |
|
P. mirabilis 1 |
0.000403 |
0.000281 |
|
P. mirabilis 2 |
0.000539 |
0.000105 |
|
P. mirabilis 3 |
0.002013 |
0.000250 |
|
P. mirabilis 4 |
0.000129 |
0.000102 |
|
P. mirabilis 5 |
0.000532 |
0.000100 |
Table 8 The folding change of treated fliL gene expression in P. vulgaris
|
Isolates |
Folding change 2˄∆∆ct |
|
|
|
Sliver (8 mg/mL) |
Titanium (10 mg/mL) |
|
P. vulgaris 1 |
0.000183 |
0.000112 |
|
P. vulgaris 2 |
0.000105 |
0.000154 |
|
P. vulgaris 3 |
0.000213 |
0.000144 |
|
P. vulgaris 4 |
0.000752 |
0.000117 |
|
P. vulgaris 5 |
0.006104 |
0.000115 |
Conclusions
Our study showed that both Ag NPs and TiO2 NPs caused swarming reduction by the down-regulation of fliL gene expression of P. mirabilis and P. vulgaris in vitro. Nanomaterials can be employed in curing many infections caused by Proteus spp. as the chance of colonization and reaching to other sites of urinary tract are reduced. We encourage using these inhibitors, unless their toxicity for human is proven, in combination with antibiotics to ensure bactericidal/bacteriostatic effect to treat risky Proteus infections.
Acknowledgments
This work was conducted by the efforts and funding of the researchers themselves and they did not receive support from any governmental or non-governmental parties. Also, we would like to thank the post-graduate M.Sc. student Muthana M. Altaay for providing bacterial isolates for the study.
References
Copyright© Tahreer Hadi Saleh, Saba Talib Hashim, Salma Nassrullah Malik, and Bahaa Abdullah Laftaah AL-Rubaii. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.