Research Article
In Vivo Genotoxicity of Gold Nanorods in Mouse Bone Marrow Compared with Cyclophosphamide
Amira M. Gamal-Eldeen 1, 2*, Mona A.M. Abo-Zeid 2, 3, Sherien M. El-Daly 1, 4, Mahmoud T. Abo-Elfadl 1, 2, Cinderella A. Fahmy 1, 2, Moustafa R. K. Ali 5, 6, Mostafa A. El-Sayed 6
1 Biochemistry Department, National Research Centre (NRC), Dokki 12622, Cairo, Egypt.
2 Cancer Biology and Genetics Laboratory, Center of Excellence for Advanced Sciences, NRC.
3 Genetics and Cytology Department, National Research Centre, Cairo, Egypt.
4 Department of Medical Biochemistry, National Research Centre, Cairo, Egypt.
5 Advanced Material Sciences and Nanotechnology Laboratory, Centre of Excellence for Advanced Sciences, NRC, Cairo, Egypt.
6 Laser Dynamics Laboratory, School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, GA 30332-0400, USA.
* Corresponding authors: E-mail: aeldeen7@yahoo.com; monaabozeid@yahoo.com
Received: Dec. 14, 2016; Accepted: Dec. 27, 2016; Published: Dec. 29, 2016.
Citation: Amira M. Gamal-Eldeen, Mona A.M. Abo-Zeid, Sherien M. El-Daly, Mahmoud T. Abo-Elfadl, Cinderella A. Fahmy, Moustafa R. K. Ali, and Mostafa A. El-Sayed, In Vivo Genotoxicity of Gold Nanorods in Mouse Bone Marrow Compared with Cyclophosphamide. Nano Biomed. Eng., 2016, 8(4): 306-314.
DOI: 10.5101/nbe.v8i4.p306-314.
Abstract
Gold nanorods (GNRs) are now under extensive investigation for biomedical applications. The in vivo genotoxic profile GNRs are not elucidated yet, therefore we investigated it in comparison with one of the most effective chemotherapeutic agents, cyclophosphamide (CP), in a mice model. PEGylated-GNRs (50 nm) were injected to Balb/C mice triple times, while CP-treated mice were treated once and the bone marrow cells (BMCs) were collected after 21 days. Chromosome aberrations, mitotic index, sister chromatid exchanges (SCEs), replicative index, micronucleus (MN) and DNA damage using comet assay were investigated. GNRs induced chromosome aberrations including- and excluding-gaps significantly at p < 0.001 and p < 0.01, respectively, however CP resulted in a higher significant increase in both types with p < 0.001. The percentage of SCEs / cell was not affected by GNRs treatment, while it was extremely significant with CP. Both mitotic activity and proliferative index were reduced dramatically with both of GNRs and CP. The recorded MN were lower in GNRs- than CP-treated mice. The percentage DNA damage, tail length and tail moment were higher in CP than GNRs. In conclusion, CP induced extreme genotoxicity more than GNRs. Both of GNRs and CP induced DNA damage. The study indicated the advantage of lower GNRs genotoxicity than that of CP. After 21 days, one injection of CP led to extreme persistent genotoxic effect more than that of multiple injections of GNRs.
Keywords: Gold-nanorods; Cyclophosphamide; DNA damage; Genotoxicity; Chromosome aberrations; Micronucleus; Comet assay
Introduction
Metallic nanoparticles (NPs) applications have gained much attention in different aspects of biomedicine, owing to their unique size-dependent properties [1]. Although bulk gold is an inert metal, gold NPs (GNPs) have been considered as the most studied and well suited NPs in the biomedical applications such as cancer treatment, drug delivery and biological imaging [2]. GNPs are produced in different shapes including nanospheres, nanorods (GNRs), nanoshells, and nanocages with different sizes ranging from few nanometers to tens of nanometer [3]. The cell/NPs interaction necessitates a precise health risk assessment of NPs effect on cells. However, most of the studies focused on the evaluating the cellular cytotoxic effect of GNPs [3-5] and few studies depicting the potential genetic alterations (genotoxicity) that can be caused by Au-NPs [6-9]. Assessment of GNPs genotoxicity depends on several factors that can influence the toxicological responses: a. GNPs physio-chemical characteristic (shape, size, and surface charge, b. the type of the stabilizing coating agent, c. the dose and the incubation time, d. the type of cells applied to, and e. the type of assays used for assessment [4, 5, 9]. For all of these factors, conflicting results about the genotoxic effect of GNPs were raised. For example Li et al. [9] reported the presence of DNA strand breaks and chromosome breaks following to short-term treatment of GNP to lung fibroblast cells, where this observed genotoxic effect, interestingly, persisted even after cell population doubling. Paino et al. [6] reported that a single short-term treatment of hepatocellular carcinoma cells (HepG2) and peripheral blood mononuclear cells (PBMC) with different concentrations of GNPs-citrate or GNPs-polyamidoamine dendrimers showed a genotoxic effect even with low concentrations of GNPs, moreover they observed that the DNA damage index was less for PBMC comparing to HepG2 upon exposure to GNPs, a finding that indicated cell specificity. According to these studies the observed DNA damage is caused either as a result of oxidative stress or thought the direct interaction of GNPs with DNA. On the other hand, instillation of colloid GNPs with different particle size (2, 20 or 200 nm) into the trachea of Wistar rats did not show a distinct DNA damage or initiation of micronucleated- polychromatic erythrocytes after 3 days of GNPs administration [10]. It is important to point out that all previous reports focused on investigating the GNPs genotoxicity after a short-term treatment period (few to 72 h), using a single administration dose and mostly applied on culture cells. Another lack in these studies was their relay on a single genotoxicity analysis (comet assay or chromosomal aberration), which is not enough to cover all forms of DNA damage nor provides an overall conclusion. As a successful strategy to treat cancer, DNA damaging agents are commonly used as anti-cancer drugs due to their high therapeutic efficacy. Since DNA replication is an elemental cell cycle phase, therefore DNA is an important target of many cancer therapies. Cytotoxic drugs that are widely used to treat cancerous patients are regularly leading to a high DNA damage, which in turn triggers cell cycle checkpoints, cell cycle arrest and subsequently cell death [11]. Cyclophosphamide (CP) is one of the nitrogen mustards that are most commonly used today as chemotherapeutic and immunosuppressant drugs in the market [12]. Alkylating agents including nitrogen mustards, CP, were reported to damage DNA by forming guanine-adenine and guanine-guanine inter-strand crosslinks are widely used as anti-cancer agents. [12]. In the present study, we were interested in evaluating the potential genotoxic effects of multiple doses of GNRs in a comparative analysis with one dose of CP, as a positive control drug, which has been extensively reported as a genotoxic anti-cancer drug through its ability to induce dominant DNA damages, as a result of free radicals and reactive oxygen species (ROS) generation. Our results may direct scientists to know the best strategies to use GNRs in biomedical approaches.
Material and Methods
Material
PEGylated-GNRs (50 nm, aspect ratio 5.0) were synthesized according to the modified seed-mediated protocol of Nikoobakht and El-Sayed [13]. The GNRs characterization was carried out by the absorption spectrum measuring using a Jasco UV-vis-near-infrared spectrophotometer, V-630 and the rods size / morphology was examined by transmission electron microscope (TEM) type JEOL-JSGM T1230, as shown in Fig. 1. Male wild-type BALB/C mice (5-6 weeks-old mice; 18-20 g; Theodor Bilharz institute, Cairo, Egypt) were maintained in a temperature controlled environment at 24°C with a 12 h light/dark cycle, and were provided with drinking water and feed ad libitum. Animal experiments were carried out according to the guidelines for the animal care statements of the Ethical Committee, National Research Centre, Cairo, Egypt. We followed the international guidelines in humane care of animals. The mice were subdivided into groups (20 mice/group): The first group was injected intravenously (IV) with GNRs (180 µg in saline/kg b. wt/week), the second group (positive control) was injected intraperitoneally (IP) with CP (20 mg/kg b. wt.) and the third group was IV-injected with saline and then bone marrow cells (BMCs) were collected from both femurs after 21 days.
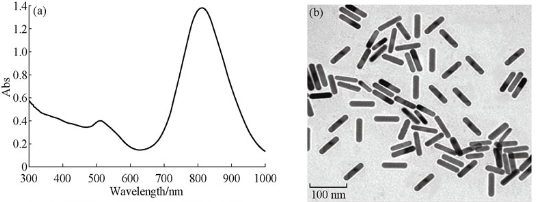
Fig. 1 (a) The electronic absorption spectrum of GNR shows two resonances with absorption maxima at 530 and 810 nm, and (b) the TEM analysis of GNRs.
Chromosome aberrations
The chromosomes preparation from BMCs were carried out according to the method of Yosida and Amano [14]. In brief, BMCs were incubated in hypotonic solution for 20 min at 37°C, and then centrifuged. BMCs bullet was re-suspended in a fixative (methanol/acetic acid) and centrifuged. The re-suspended cells in fixative were spread by dropping onto frozen slides. After drying, the slides were stained by 10% Giemsa for 40 min, washed, and air dried. Fifty well-spread metaphases were analyzed per mouse. Metaphases with gaps, chromosome or chromatid breakage, fragments, deletions and numerical aberrations (polyploidy) were recorded.
Mitotic index
The prepared slides for chromosomal aberrations were used to determine the mitotic index (MI), which based on the scoring of 1000 cells for each mouse. The number of dividing cells including prophases and metaphases was recorded. The mean percentage of mitotic index was calculated according to the formula: MI = (number of dividing cells/1000 cells) *100.
Sister chromatid exchanges
5- Bromodeoxyuridine (BrdU) tablets, prepared with a Parr press, were implanted subcutaneously in mice to dissolve gradually [15]. BMCs were isolated after 24 h from treatment. Two hours before collecting samples, mice were injected with colchicine. The fluorescence-photolysis Giemsa technique was used [16]. The slides were stained with Hoechst 33258 dye. At 50°C, the slides were layered with Mcllvain’s buffer, covered with coverslips and subjected to fluorescent black-blue tube. The slides were stained with Giemsa stain solution and air dried. The frequency of sister chromatid exchange (SCE's) was recorded for each animal by counting number of SCEs in 30 cells of second division. The frequency of SCEs/cell was determined.
Replicative index
The estimation of changes in cell kinetics was obtained by determining the replicative index (RI) using slides prepared for SCEs analysis. One hundred metaphase cells per animal were analyzed and the number of first (M1), second (M2), and third (M3) divisions were recorded. The RI was calculated according to the equation: RI = (1M1 + 2M2 +3 M3) x 100 [17].
Micronucleus test
BMCs micronucleus test was carried out according to Schmid [18] to evaluate chromosomal damage in experimental animals. BMCs were flushed out gently with fetal calf serum (FCS). The cells were collected by centrifuging at 1500 rpm for 10 min at 4°C. Cell pellet was re-suspended in a drop of FCS and smears were prepared on to glass slides, air-dried, fixed by absolute methanol and stained by May-Grünwald/ Giemsa as described by Schmid [18]. For each mice group, the number of BMCs containing micronuclei (MN) was recorded to the total number of BMCs.
Comet assay
DNA damage was detected using the alkaline version of the Comet Assay as described by Schlörmann and Glei [19]. Comets were examined at 400X magnification using a fluorescent microscope with image analysis system (Perceptive Instruments, Comet assay V2). Sixty cells per slide were scored and recorded for each animal. The mean values ± SE for percentage of DNA in tail, tail length (µm) and tail moment were recorded.
Statistical analysis
The statistical analysis of chromosome aberrations was done according to Chi-square- contingency test. MI, SCEs, RI, MN and Comet assays statistics were carried out by one-way analysis of variance (ANOVA) (Tukey-Kramer Multiple Comparisons Test) using GraphPad Stat software. All data were presented as means ± SE and results were considered statistically significant when p < 0.05.
Results
After 21 days from GNRs treatment (180 µg kg b. wt./week, for three weeks), BMCs were collected from GNRs-, CP- and control groups to assess chromosome aberrations, mitotic activity, sister chromatic exchanges and replicative index, besides measure the potential of GNRs to induce DNA damage using comet and micronucleus tests. The mean percentages of chromosome abnormalities with and/or without gaps were recorded in BMCs (Table 1) after IV-treatment of GNRs and compared with negative and positive control groups. CP, positive control, is a known anticancer drug in the market, which is characterized by its potential to induce chromosome abnormalities in an extremely significant extent (p < 0.001) with or without gaps, as shown in Table 1. GNRs significantly induced chromosome aberration including (p < 0.001) and excluding gaps (p < 0.01) up to 2.26- and 2.04-folds of control, respectively, however it was obviously noticed that CP is a dramatic genotoxic drug that enhanced chromosome aberration including (p < 0.001) and excluding gaps (p < 0.001) up to 6.0- and 7.04- folds of the control, respectively (Table 1). Different types of chromosome aberrations including gaps, breaks, fragments, deletions, and tetraploid cells were recorded (Table 1, Fig. 2(a)-(c)). CP- and GNRs- treatments increased the number of chromatid breaks 3.13- and 2.88-folds (p < 0.01) from control respectively. We found that cells contained more than one aberration, multiple aberrations, were increased extremely (p < 0.001) when animals treated with CP only in comparison with control. The more chromosome abnormalities, the decreased mitotic activity of the cells were recorded with CP- and GNR- treated mice. This was indicated when the mean percentages of mitotic indexes reduced significantly (p < 0.001) down to 47.73% and 77.64% of control, respectively (Fig. 3(a)). In SCEs analysis (Fig. 2(d)-(f); Fig. 3(b)), there was a non-significant increase (p > 0.05) in the number of SCEs / cell in GNRs-treated group with a total counted number of 1466 SCEs / group compared to that of the control (977 SCEs/ group). On the other hand, CP-treated group exhibited a dramatic elevation (p < 0.001) in both of the number of SCEs / cell and the total counted SCEs number (6696 SCEs / group). In contrary, the cell proliferation (replicative index) was degenerated (p < 0.001) in both of the GNRs- and CP-treated groups (Fig. 3(c)). For MN analysis (Fig. 2(g)-(i)), both of the GNRs and CP remarkably induced DNA damage (p < 0.001) in BMCs. The MN results indicated a significant increase (p < 0.01) in the mean percentages of MN in GNRs-treated group with a total counted number of 274 MN / group compared to that of the control (122 MN / group), while the treatment with CP led to a 4.5 fold elevation (p < 0.001) in the mean percentages of MN, and the total counted MN number reached to 542 MN/ group (Fig. 3(d)). As shown in Table 2, comet assay was used as a sensitive analysis to study the damaged or fragmented DNA per one cell in isolated BMCs from GNRs- and CP- treated mice in comparison with control mice. The microscopic analysis (Fig. 2(j)-(l)) revealed that GNRs and CP treatments led to a noticeable DNA damage (p < 0.001) in BMCs as concluded from the high frequency of tail moments resulted from the long tail length of comets (3.5 and 4.8 – folds of control, respectively) and high percentages of DNA damage (5.02 and 5.28 – folds of control, respectively) (Table 2).
Table 1 Number and mean percentages of chromosome aberrations induced in different mice groups treated with CP or GNRs.
|
|
Abnormal metaphases |
No. of different types of aberrations |
||||||||
|
Including gaps |
Excluding gaps |
Gaps |
Br |
F |
Del |
MA |
Tp |
|||
|
No. |
Mean (%) |
No. |
Mean (%) |
|||||||
|
Control |
35 |
4.38 ± 0.20 |
26 |
3.25 ± 0.27 |
9 |
8 |
12 |
3 |
0 |
3
|
|
CP |
210 *** |
26.25 ± 0.94 |
183 *** |
22.87 ± 1.11 |
27 **
|
25 ** |
19 |
8 |
120 *** |
11*
|
|
GNRs |
79 *** |
9.88 ± 0.51 |
53 ** |
6.63 ± 0.40 |
26 ** |
23 ** |
19 |
10 |
0 |
1 |
The total number of scored metaphases is 800 (n = 16 mice/group); *p < 0.05; **p < 0.01; *** p < 0.001: Significance compared to Control; Chi-square- contingency test using GraphPad Stat; F: Fragments, Br: Breaks, Del: Deletions, MA: Multiple Aberrations, Tp.: Tetraploidy.
Table 2 Comet analysis (% of DNA damage, Comet tail length, and Comet tail moment) in different mice groups treated with CP or GNRs.
|
|
DNA damage (%, mean ±SE) |
Comet tail length (µm, mean ±SE) |
Comet tail moment (mean ±SE) |
|
Control |
3.162 ± 0.252 |
3.552 ± 0.135 |
0.274 ± 0.049 |
|
CP |
16.706 ± 1.679 *** |
17.048 ± 1.244 *** |
4.417 ± 0.652 *** |
|
GNRs |
15.872 ± 1.343 *** |
12.522 ± 1.296 *** |
3.171 ± 0.468 *** |
Total number of scored cells is 60 cells/mouse (n = 16 mice/group); ***p < 0.001: Significance compared to control; One-way Analysis of Variance (ANOVA)-Tukey-Kramer Multiple Comparisons Test.
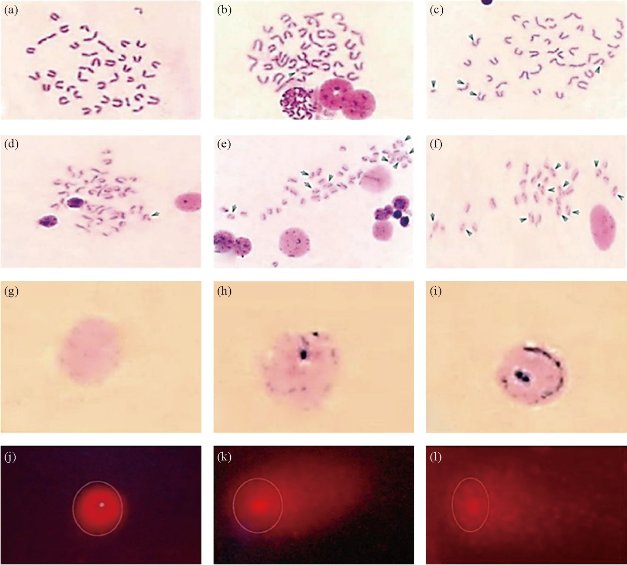
Fig. 2 Representative photo panels for microscopic cytogenetic profiles of BMCs for chromosome aberrations ((a) normal metaphase, (b) gap, and (c) MA including fragment, breaks and deletions; x1000), SCEs ((d) few numbers of SCEs, (e) moderate numbers of SCEs , and (f) high numbers of SCEs; x1000), MN ((g) normal PCEs, (h) and (i) PCEs with micronuclei; x1000), and comet assay ((j) intact cell, (k) highly damaged cell, and (l) extremely damaged cell; x1000).
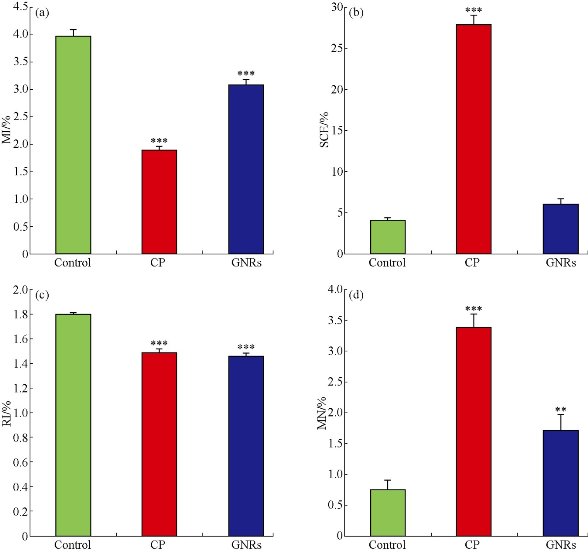
Fig. 3 Mean percentages of MI, SCEs/cell, RI and MN in BMCs of GNRs- and CP- treated mice in comparison with control. (a) MI: The number of divided cells (prophases and metaphases) to total number of scored cells was recorded in 1000 cells/mouse; (b) SCEs: The number of scored cells was 30 cells/mouse; (c) RI: The number of examined cells was 100 cells/mouse; and (d) MN: The number of examined cells was 1000 cells / mouse; n = 16; **p < 0.01; ***p < 0.001: significance compared to Control; One-way Analysis of Variance (ANOVA) test using GraphPad Stat.
Discussion
This study would be considered as one of the premier studies carried out in vivo investigation of cytogenetic effects of GNRs, particularly. CP is a clastogenic agent for various animal species and is a mutagenic agent usually used as positive control in in vivo tests of short duration. Chorvatovicová and Sandula [20] recommended CP to be used in chromosome aberration, sister chromatid exchanges and MN formation in vitro and in vivo experiments. It is characterized by its potential to induce chromosome abnormalities with or without gaps. As a widely used anticancer drug with effective cytotoxic/genotoxic properties, we used CP to compare with the genotoxic properties of GNRs. Evaluating the cytotoxicity and/or genotoxicity potential of GNRs in comparison with the chemotherapeutic agent CP, in mice BMCs, was assessed by different cytogenetic analyses characterized by their most reliable, sensitive and efficient indications. Our findings revealed that, although both of CP and GNRs are inducers of chromosome abnormalities, but GNRs induced abnormalities to far lower extent than CP, where in abnormalities including gaps CP was 2.66-folds > GNRs and in abnormalities excluding gaps CP was 3.45-folds > GNRs. However the ratio of chromosomes and/or chromatid breaks, that were remarkably higher than control in both of CP and GNRs, were in similar percentages in both. It worth to be mentioned, the complete absence of multiple aberrations in GNRs, while CP resulted in a dramatic enhanced multiple aberrations (n = 120). Furthermore, the number of SCEs/cell was increased in GNRs-treated mice to 1.5-folds of the control, and increased extremely in CP-treated mice up to 6.86-folds of the control. Previous reports for CP and its analogues demonstrated its cyto/genotoxic effects [21-23]. GNRs results are in agreement with the studies of Li et al. [9] who demonstrated the genotoxicity of another structure, sphere GNPs, which induced chromosome aberrations per cell in MRC-5 lung fibroblasts to up to 4.0-folds of control. They also found that most of the observed aberrations were chromosomal breaks with the majority of undetectable telomeres, as detected by FISH. They also reported that these abnormalities persisted even after one population doubling. In the light of this report and in comparison to our results, although GNRs were injected to mice triple times before collecting BMCs after 21 days, while CP was injected only once, but CP exhibited extreme genotoxic effect more than that of multiple injections of GNRs. It can be also suggested from the comparison of our results with this previous report that there is no structure influence on the chromosomal abnormalities, since both of gold nano-spheres and -rods resulted in chromosomal aberrations. The mitotic and replicative indexes are other useful endpoints for assessing the reproductive competence of cells. Not only they reflect the efficacy of cells to make DNA synthesis, but also indicate their potential to progress through G2, and reveal chromatin condensation, which consequently form chromosomes [24]. In our study we observed that the more chromosome abnormalities and SCEs; the decreased mitotic and replicative activities of the cells with GNRs- or CP-treated mice and that cell proliferation was reduced remarkably in both mice treated groups. This reduction in cell proliferation may be due to the potential of GNRs to arrest cell cycle at G1/S phase as CP. Alkan et al. [25] demonstrated that CP (10 –25mM) reduced cell proliferation, as evaluated by MTT and LDH test, and caused accumulation of canine mammary tumor cells in S phase, and observed extreme reduction in G0/G1 phase cells. Connor et al. [26], demonstrated that spheres GNPs (diameter 4-18 nm, covered with citrate, cysteine, glucose, biotin, and cetyltrimethylammonium bromide) were non-toxic on human leukemia K562 cells. Singh et al. [27] reported that low concentration of oleic acid- sophorolipid GNPs up to 50 µM did not induce cyto/genotoxicity in HepG2 cells. In contrary, other researches such as Patra et al. [28] reported that 13 nm citrate-coated GNPs exhibited toxicity against human carcinoma lung cells 20-120 nM and Paino et al. [6] mentioned that both PAMAM- and citrate-coated GNPs induced cytotoxicity in HepG2 cells or PBMC, using the MTT assay. They referred GNPs cyto/genotoxicity to their small size and nanometric dimensions, which could diffuse through cellular membrane and thus uptake by cells [29, 30]. Recently, we reported the potential of GNPs to reduce the cell proliferation and to induce apoptosis in human blood lymphocytes in vitro [31, 32]. In these studies, we found that different shapes of GNPs (big and small rods, sphere and semi-cube shapes (0.05-1.0 μg/ml) initiated duplication of CASP3, CASP7, CASP9 and TP53 genes signals with I-FISH, which are specific for apoptotic pathway, and reduced signals of TNF and CRP genes, which are used as markers for inflammatory response and necrosis. MN test is one of the simplest and sensitive screenings modality to assess the structural chromosome damages induced by clastogenic agents in mice BMCs [33, 34]. Micronuclei appear in the cytoplasm as small condensed chromatin bodies fragmented from acentric chromosomes or whole chromosomes [35]. Comet assay, single cell gel electrophoresis, is a powerful tool to detect genotoxicity, DNA damage/repair and biomonitoring for clastogenic or bio-materials in eukaryotes [27]. We found that the CP-induced MN percentages were double fold higher than GNRs-treated mice. On the other hand, the ratio of tail moment, tail length and percentage of DNA, in comet assay, were extremely high and similar in in both of GNRs- and CP- treated mice. Previous works recoded the mutagenic effects of CP, and its potential to induce micronuclei and DNA damage [21-23]. On the other hand, other structure such as sphere GNPs potentially induced DNA damage as previously reported [6, 9, 31, 36]. This DNA damage could be correlated to the small size of GNPs, which give them the ability to diffuse freely through nuclear pores to become in contact with DNA [26, 37-39]. The direct contact of GNPs with DNA may trigger DNA damage through apoptosis [32, 39, 40], and dysregulate DNA repair genes [41], which in consequence lead to persistence of DNA damage. The high efficacy of GNPs to kill cancer cells more efficiently than normal cells was observed in the studies of Paino et al. [6], who stated that GNPs-sodium citrate or GNPs-PAMAM induced in vitro genotoxicity/cytotoxicity in HepG2 cancer cells and PBMC primary cells even with low concentrations and that both induced DNA damages in HepG2 cells higher than in PBMC. In conclusion GNRs induced genotoxicity as concluded from chromosome aberrations, SCEs and MN but to a lower extent than the chemotharpeutic drug CP. Both of GNRs and CP similarly induced sever DNA damage as evaluated with comet assay. The study indicated the advantage of lower genotoxicity of GNRs than CP accompanied with reduced mitotic activity and replicative index. One injection of CP led to extreme genotoxicity more than that of multiple injections of GNRs.
Acknowledgements
This work was funded by Misr el Kheir Foundation and The National Research Centre (NRC), Cairo, Egypt. We would like to thank Prof. Ali Shabaka, Spectroscopy Department, National Research Centre, Cairo, Egypt, for his guidance through the project.
Conflict of Interest
Authors declare no conflict of interest.
References
Copyright© 2016 Amira M. Gamal-Eldeen, Mona A.M. Abo-Zeid, Sherien M. El-Daly, Mahmoud T. Abo-Elfadl, Cinderella A. Fahmy, Moustafa R. K. Ali, and Mostafa A. El-Sayed. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.