Review
Superoxide Dismutase, a Potential Theranostics against Oxidative Stress Caused by Nanomaterials
Chao Li, Daxiang Cui *
Department of Bio-Nano Science and Engineering, Key Laboratory for Thin Film and Micro fabrication Technology of Ministry of Education, National Key Laboratory of Micro /Nano Fabrication Technology, Research Institute of Micro/Nano Science and Technology, Shanghai Jiao Tong University, Shanghai, 200240, P.R. China.
*Corresponding author. Email: dxcui@sjtu.edu.cn Tel/ Fax: 008621-34206886
Citation: C. Li et al. Superoxide dismutase, a potential theranostics against oxidative stress caused by nanomaterials. Nano Biomed. Eng. 2012, 4(4),195-206.
DOI: 10.5101/nbe.v4i4.p195-206.
Abstract
Orthopaedic infections represent one of the major causes of implant failure. Systemic treatment is limited due to dosing, side-effects, patient compliance, treatment length and resistant bacteria. The choice of antibiotic incorporation method has been the subject of many investigations and, nowadays, various vehicles for local drug delivery have been studied. In this work, a novel ciprofloxacin loaded chitosan nanoparticles coating system onto titanium surface has been developed and characterized. The antibiotic release capability of this system and its ability to inhibit the in vitro growth of two of the most common pathogens causing orthopaedic implant-related infections, Staphylococcus aureus and Pseudomonas aeruginosa, have been evaluated. Preliminary biocompatibility data arising from MG63 osteoblast-like cells seeding on the ciprofloxacin-loaded systems have also been discussed. The investigated system represents a promising candidate in view of the development of new antibiotic carriers in situ for preventing titanium implant-associated infections.
Keywords: Superoxide dismutase, Nanomaterial, Nanocarrier, Drug delivery system
1. Introduction
With the fast development of nanotechnology, a lot of nanomaterials are continuously fabricated, and are actively explored their applications in pharmaceuticals, cosmetics, biomedical engineering, nanoelectronics, etc. The emerging nano-industries may create enormous riches, but the risk of nanomaterials and nanotechnology on environment and human healthcare is still not clarified well. Up to date, some data show that nanomaterials can result in potential risk. For example, all the nanomaterials such as carbon nanotubes, graphene, sliver nanoparticles, gold nanoparticles, etc. can cause serious cellular oxidative stress, finally result in cell apoptosis or death [1]. How to prevent the damages from the oxidative stress caused by nanomaterials have become our concerns. Superoxide dismutase (SOD) is one kind of enzyme, also called as liver protein or orgotein, widely distributed in animal, plant and microorganism. SOD performs some special functions inside the live cells such as combating with the reactive oxygen species (ROS), interfering with the injured made by the hydrogen peroxide (H2O2), and repairing the damage induced by these ROS. SOD is not only the primary material that could eliminate the effects made by free radical species, and also prevent and hold back the effete and eclipse of living organism. In the past decade, a great deal of investigations demonstrate that SOD is a foremost protective bioenzyme, it can resist the superoxide free radicals effectively, extend life span, and adjust the metabolism of live cells, enhance the organism immune function. Therefore, SOD maybe one kind of excellent potential theranostics against oxidization in vivo, owns great potential application in therapy of oxidative stress-induced damage made by nanomaterials. Although SOD exhibits great application prospects in clinical anti-oxidization therapy, SOD based novel pharmaceutical development meets with some difficulties. For example, how to highly efficiently deliver SOD into therapeutic target locus in vivo, how to make endosome or lysomes filled with SOD effectively release SOD into cytoplasm, all these problems have become great challenges. Herein we review the main advances of SOD and its high efficient delivery systems, and explore the issue challenges, and prospects with the aim of developing novel nanoscale theranostics against oxidative stressinduced damages caused by nanomaterials.
2. The structure and function studies of SOD
SOD is one kind of biological active substance derived from the live organisms, and it can clean up the harmful free radical species produced in the metabolic process, constant supplement of SOD has special preventive effects for treatment of oxidative stress.
2.1 History and function of SOD
In 1938, Mann and Keilin extracted one kind of protein containing copper from the bovine erythrocyte [2], and in 1953, Keilin extracted the hepatic copper protein from vituline and whale liver [3]. In 1968, Dr. I. Fridovich and his student McCoard. J. M in Dude university nominated the copper protein derived from blood, liver and brain as superoxide dismutase according to SOD catalytic activity [4]. SOD is widely distributed in live organisms. It may come from body of animal to plant vegetation, and from the whole body to local cells. SOD was found from bovine erythrocyte at first, which was usually extracted from the blood cell of cattle or some other animals with a very costly consume. At the same time, because of SOD’s antigenic heterogeneity, inclinable to denaturalization in normal temperature, the virus in blood products and other hazard factors could lead to cross-infection, WHO declared that the utility of SOD derived from animal fountainhead materials should be stopped immediately. European Union had laid a ban to prohibit using the SOD from wild animals. Up to date, many techniques have been developed to extract SOD from plant or the fermentation of modified bacteria transferred with plasmid. SOD catalyzes the conversion of superoxide to oxygen and hydrogen peroxide. This biochemical reaction helps to break down potential harmful oxygen molecules in cells, which might prevent ROS damage to tissues. The biochemical reaction can be summarized as the following formulate: M3++ O2−→M2++O2 M2++ O2−+2H+→M3++H2O2 Some cellular processes produce O2−, called superoxide anion free radical, which is one of the intermediate products in natural physiological reaction of living cells. This active oxygen has extra potential ability of oxidation and also is the important factor of biological oxygen toxicity. Except the O2−, the hydroxyl radical, superoxide radical, alkoxyl radical, polyunsaturated fatty acid radical, semiquinone radical, singlet oxygen 1O2−, hydrogen peroxide (H2O2), nitrite (NO2−) and nitric oxide (NO). Although these radicals have not the single electron, they have the comparative ability of oxidation and toxicity to living cells, oxidizing and degrading biological important molecules such as lipids and proteins, they are called as the active oxygen consequently. SOD can transform these oxide radicals into hydrogen peroxide, although the hydrogen peroxide has biohazard activity, it can be quickly decompounded into such harmless and safe products as H2O and O2 by high activity catalyse and peroxidase that widely distributed in living cells. In this way, a complete anti-oxide chain is composed by SOD, CAT and POD in live organism.
2.2. The category and biological properties of SOD
SOD is widely distributed in our environment and has been reported in many species, from prokaryocyte bacteria to eukaryote cells, as well as microorganisms, plants and animals. SOD is classified as follows, based on the requirement of the metal species at the active site [5]: (A): Copper- and zinc-containing superoxide dismutase (CuZnSOD) [4]. (B): Iron-containing superoxide dismutase (FeSOD) [4] (C): Manganese containing superoxide dismutase (MnSOD) [4] (D): Nickel-containing superoxide dismutase (NiSOD) [6] (E): Iron- and zinc-containing superoxide dismutase (FeZnSOD) [6] (F): Cobalt- and zinc-containing superoxide dismutase (CoZnSOD) [7] Furthermore, some SOD’s biological properties were well summarized in Table 1.
Table 1. Biological property of three types of SOD
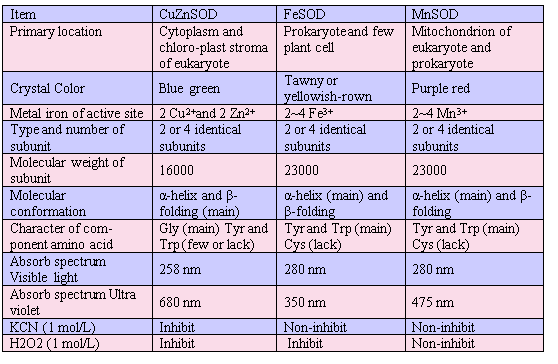
2.3 Structure and function of CuZn SOD
CuZnSODs have been extracted from a wide range of organisms including yeast, spinach, gallinaceous liver and bovine blood. In all these cases, a homodimeric enzyme is obtained, with a molecular weight of 3.1~3.3 kd and containing one Cu2+ (direct related with enzyme activity) and one Zn2+ (related with structural stability) per subunit [8,9]. Each subunit is composited with 150 amino acids and one bio-active center. Fig. 1 is the crystal structure model of bio-active site [9]. The sequence of CuZnSOD derived from many microorganisms, plants, fishes and mammals, demonstrated that the CuZnSOD displayed a high evolutionary conservation and sequence homology[10]. The pure crystal of CuZnSOD presents a cyan color and the maximum absorption value locates at 258 nm.
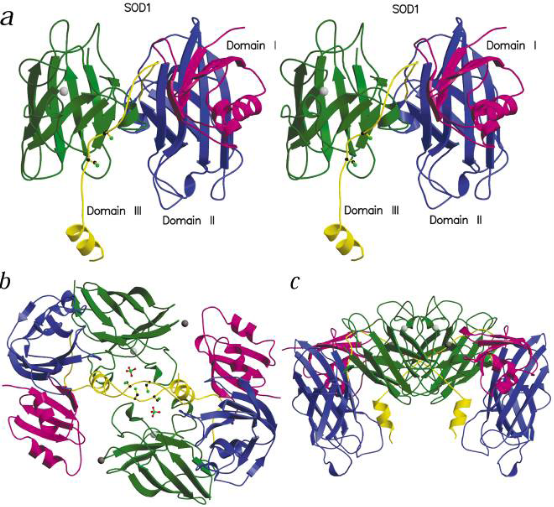
Fig. 1 The model of recombined human CuZnSOD (a: monomer, b: the top view of dimerization, c: side view of dimerization). Copyright permission from ref. 8.
2.3.1 Property of two dimension (2D) structure of the CuZnSOD
1) The amino acid remnant sequences of the assistant subunits attached with metal (Cu and Zn) active site are identical with the vicinal amino acid sequences that composite the inner disulfide bond (Fig. 2). 2) CuZnSOD is abundant in Glycin and distribute uniformly in the whole sequence. For example, there are 25 Glycin residues in 151 whole sequences in human CuZnSOD of erythrocyte; it is 1/6 of total amino acids. 3) There is a high conserved sequence in the C-terminal end and Arginines in this area are closely related with the activity of the CuZnSOD enzyme. 4) There is a high variable sequence composited with 23~35 amino acid remnants in the N-terminal end of CuZnSOD; it may correlate with the immunological properties of the CuZnSOD enzyme.
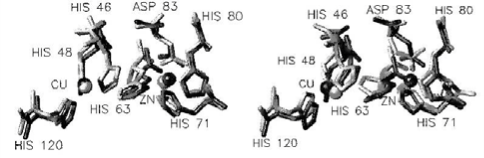
Fig. 2 The amino acid distribution of the active site of human SOD (left: deoxidization type, right: oxidation type). Copyright permission from ref. 9.
2.3.2 Property of three dimension (3D) structure of the CuZnSOD
The high resolution X-ray diffraction in 1.15 Å or 1.65 Å to bovine CuZnSOD crystal analysis and some other examination illustrated the 3D structure of the CuZnSOD has the following characteristics: 1) The fold of each subunit, its volume and surface are about 20600Å3 and 12390 Å2, respectively. The two subunits combined by dehydrophilic covalent bond, the interface between the subunits is extremely stable due to the 1038Å2 connected area [11]. 2) The disulphide bridge between Cys55 and Cys144 is a conserved feature in all structure of CuZnSOD and contributes significantly to the stability of the enzyme. 3) In the whole structure of each subunit of CuZnSOD, α helix is about 5% and β helix is about 45%~, an eightstranded antiparallel amino chain constitute a “greekkey” and three long, external loop regions connected to the β barrel. Two of them together with a section of the β barrel form the walls of channel, the so-called “active site cavity” leading from the enzyme surface to the enzymatic active site. The third loop provides the greekkey connection across the β barrel (Fig. 1) [12]. 4) The distance between the double subunits is 34 Å, meanwhile, in each monomer of CuZnSOD, the distance of the iron center between Cu2+ and Zn2+ is merely 6 Å. The closest amino acids to Cu2+ is 4 Histidins (His118, His46, His44, His61) and a molecule of water, but to Zn2+, the closest amino acid is Asp81, His78 and His61. The His61 is the primary composition of the Imidazole Bridge between Cu2+ and Zn2+ [12]. 5) Use CuZnSOD of the human erythrocyte as an example, its active center is an ellipse barrel and its length, width and depth is 15 Å, 9 Å and 6 Å respectively. Generally, in one broad of the barrel is Thr135, Gly136, Ala138 and Gly139, the other broad is composed by Gly59, Pro60, His61, Phe62 and Asp63. Lys134 and Arg141 composed the side edges of the barrel. Actually, the entire active center is homology for the CuZnSOD molecule divided from different species.
2.4 Structure and function of MnSOD
The manganese-dependent enzyme (MnSOD) was found in mitochondrion in eukaryotes, prokaryotes and protists, the amino acid sequences composed the active center are highly conserved [13]. The structure of the human MnSOD monomer shows very different from the CuZnSOD. The MnSODs have tow domains: one formed by two long α helices and one formed by five short α helices and two β sheets. The metal ion is coordinated by three His and one Asp [14]. The 3D structure of MnSOD from Escherichia coli has been determined by X-ray crystal-lography at 2.1Å resolution [13]. The protein crystallizes with tow homodimers in the asymmertric unit, and a model comprising 6528 protein atoms (residues 1-205 of all four monomers), four manganese ions and 415 water molecules has been refined to an R factor of 0.188 (Rfree 0.218). The Mn centers are 5-coordinate, trigonal bipyramidal, with His26 and a solvent molecule, as apical ligands. The coordinated solvent molecule is linked to a network of hydrogen bonds involving the non-coordinated carboxylate oxygen of Asp167 and a conserved glutamine residue, Gln146. The MnSOD dimmer is notable for the way in which the two active sites are interconnected and a “bridge” comprising His171 of one monomer and Glu170 of the other offers a route for inter-site communication [15]. Fig. 3 is the amino acid distribution and the 3D structure of human MnSOD.
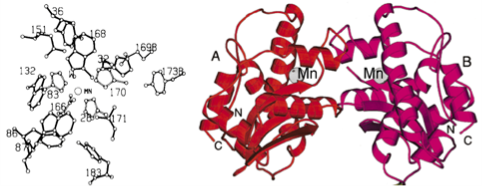
Fig. 3 The overall structure of a MnSOD subunit. (left: a amino acid distribution in active site of MnSOD. Right: Stereodiagram of tow domains structure of MnSOD. Copyright permission from ref. 16 and 19.
2.5 Structure and function of FeSOD
FeSOD was purified from Aquifex pyrophilus firstly in 1992 called Ap SOD [17]. The Ap SOD is a tetrameric enzyme and extremely stable against heat and chemical inactivation [18]; it maintains 70% of its activity after heating at 100 oC for 60 min. In the presence of 1% (w/ v) SDS, about 70% of Ap SOD retains its activity after heating at 80 oC for 60 min [19]. As shown in Fig. 4, the structure of Ap SOD can be divided into two domains. The N-terminal domain consists of two long antiparallel α-helices. The C-terminal domain contains a central β-sheet formed by three antiparallel β-strains and five α-helices surrounding it. The Fe3+ is liganded by two residues from N-terminal helices and tow residues from the loops in the C-treminal α+β domain, and forms a site for substrate binding and catalysis. In general, Fe- and Mn-containing SODs are more closely related structurally and are considered as members of one family [16]. However, the Fe3+ content gradually decreased (to 45%) during storage, suggesting that the metal ion is not tightly associated with Ap SOD. The amino acid sequences of the metal-binding site of all SODs are well conserved. In Ap SOD, iron is liganded by His27 and His81 from helix α1 and α2, respectively, Asp163 and His167 from L5, and a water molecule with distorted trigonal bipyramidal geometry; three atoms, NE2 of His81, OD2 of Asp163 and NE2 of His167, form a trigonal basal plane, and NE2 of His27 and a solvent molecule fill the two axial positions in the trigonal bipyramid [19]. The most noticeable structural difference in the active site regions between FeSODs and MnSODs is a lignad that interacts with a solvent molecule bound to metal; histidine is coordinated to a Fe-bound solvent in Ap SOD (His148). Such a difference implying that these residues may play a critical role in the metal selection [19].
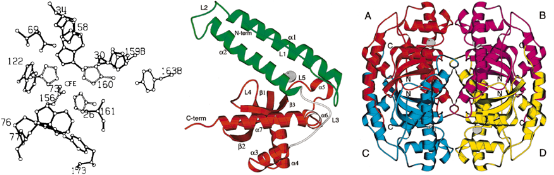
Fig. 4 The overall structure of an Ap SOD (FeSOD) subunit. (left: a amino acid distribution in active site of FeSOD; middle: Ribbon diagram of a FeSOD monomer. Right: Stereodiagram of the quaternary structure of FeSOD. Copyright permission from ref. 16 and 19.
3. Nanocarriers for intracellular SOD delivery
How to deliver efficiently SOD into local tissues for SOD's anti-oxidative function has become a big challenge. So far, nanoscale delivery systems have achieved big advances. How to design and fabricate delivery system suitable for SOD have become our concerns. In the past decades, many techniques have been developed to deliver active proteins to specific cells or organs. For example, lipid-based nanocarriers, polymeric nanocarriers, inorganic nanocarries and protein-mediated nanocarriers etc. Delivering active forms of proteins to living organisms is an important goal in many medical and biological applications, including cancer therapy, vaccination, regenerative medicine, treating loss-of-function genetic diseases and imaging. SOD is a macromolecule protein with catalytic activity. The extracellular SOD is very difficult to enter intracellular position, and eradicate the superoxide radicals for the large size, varying surface charges and fragile tertiary structures of SOD. When the natural barrier of living cells is broken by the extracellular SOD and administered into serum, the SOD enzyme may suffer from serum instability and can be rapidly degraded or inactivated. Moreover, even the SOD is made into oral agents or skin liniment, for the short half life period (15 min) of native SOD molecule and relatively sensitive to the outer chemical and physical factors, such as temperature and solution pH, it is difficult to exert its bioactivity effectively and entirely. To modify SOD molecule and enclose with nanomaterials via nanotechnology, not only could enlarge its half life efficiently, enhance its biostability in vitro or in vivo, but also leads to an improved vehicle that could deliver active SOD molecule to the target cells or organs. Fig. 5 is a schematic process of a typical endical endocytic pathway for delivery vehicles with protein cargoes, most importantly, to success enter the special site of target cells such as the plasma or the nucleus, the vehicle often needs to help the protein cargo in endolysosomal escape. To date, the most commonly used application for intracellular SOD delivery is target protein to protein transduction domains (PTDs) or cell-penetrating peptides (CPPs). Despite many practical advantages of the SOD transduction technology, the main concern of this method is the inefficient escape from the endosome to cytosol, leading to CPP-tagged cargoes sequestered in intracellular vesicles [20,22]. In the last few decades, nanocarrier-based intracellular protein delivery approaches have generated considerable interest and several promising strategies have been developed. As summarized in Fig. 6, these nanoscale carriers include lipid-containing colloidal systems such as liposomes and solid lipid nanoparticles, polymeric nanocarriers, inorganic nanoparticles/nanotubes and protein-based carriers. Target protein cargoes can be loaded into various nanocarriers using different strategies, including direct conjugation via either chemical modification or physical adsorption and covalent/ noncovalent encapsulation encapsulation [23]. One of the key functions of the nanocarriers is to increase the penetration of the delivered protein by concealing antigenic and immunogenic epitopes and attenuating receptor-mediated uptake by the reticuloendothelial system (RES) [24]. Furthermore, nanocarriers can prevent protein proteolysis and can increase the size of the delivered cargo in vivo, thus reducing renal filtration. The high surface area to volume ratio of nanocarriers also leads to improved pharmacokinetics and biodistribution of payload [25-27]. Another crucial feature of the nanocarrier-based delivery system is the increased flexibility of tailoring its physical characteristics such as size, surface charges and displayed ligands can be customized to facilitate cell penetration and endolysosomal escape, as well as to optimize bio-stability, targeting specificity and cargo release kinetics [28].
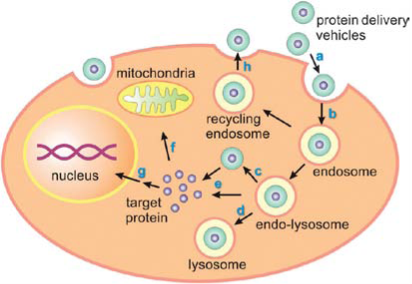
Fig. 5 Schematic process of a typical endocytic pathway for delivery vehicles with protein cargoes. Copyright permission from ref. 21.
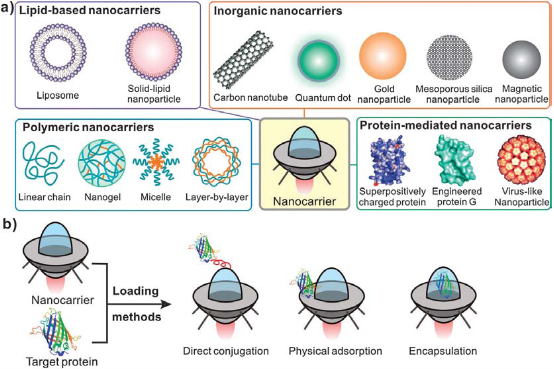
Fig. 6 Schematic of various types of nanocarriers used for delivery; b: Schematic of three main loading methods for preparing protein/nanocarrier composites. Different nanocarriers are represented as a “space shuttle”. Copyright permission from ref. 21.
3.1 Lipid-based SOD nanocarriers.
Liposomes are bilayered vesicles assembled from amphiphilic building blocks, such as lipids or phospholipids. The size of liposomes can range from 20nm to several microns. Liposome can adhere to plasma membranes and enter the cell via endocytosis or liposomecell fusion [26]. Different liposome formulations have been explored as delivery vehicles for various hydrophilic or hydrophobic compounds [25]. Lenormand and his coworkers utilized liposomes for intracellular delivery of therapeutic membrane proteins [29]. The voltage-dependent anionic channel (VDAC) and the pro-apoptotic Bak were assembled into the lipid bilayer of liposomes, forming proteoliposome particles. They demonstrated that the internalization of integrated protein/liposomes into living cells induced apoptosis by release of cytochrome C and activation of caspases. Recently, several commercial lipid-based reagents have been utilized for protein delivery and enhanced the development of lipid-based nanocarriers in biological and medical applications.
3.2 Polymeric SOD nanocarriers
To date, several SOD nanocarriers have been developed and a few of them have received the Food and Drug Administration (FDA) approval for clinical use. Generally, the nanocarriers can be constructed as follow:
3.2.1 Physical adsorption and interaction
Commonly, the chemical modification including the covalent or noncovalent conjugation may lead to deleterious effects on the activities of the cargo. The alternative method without covalent modification is the self-assembly between the protein and the nanocarrier. The spontaneous assembly is typically facilitated by physical adsorption, which is driven by electrostatic forces for van deer Waals interactions. Zimmerman and coworkers demonstrated effective intracellular delivery of copper/zinc superoxide dismutase (CuZnSOD), which can savenge O2- intermediates resulting from aberrant AngII signaling in the central nervous system in a multitude of cardiovascular diseases [30]. “Nanozymes” were created by complexing CuZnSOD with a copolymer of PEI and PEG to create polyion complexes. When the nanozymes were delivered to mouse catecholaminergic CATH neurons, the increase in O2- intermediates caused by AngII was significantly inhibited compared to untreated neurons or neurons treated with native CuZnSOD. When CuZnSOD nanozymes were injected in vivo using an intracarotid injection into rabbits along with intracerebroventricular-delivered without any toxic effects. In comparison, intracarotid injection of free CuZnSOD or PEI-PEG copolymer did not exhibit inhibition of AngII responses (Fig. 7).
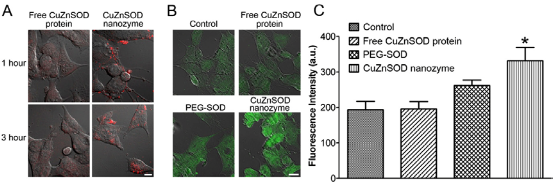
Fig. 7 CuZnSOD nanozyme penetrates CATH. A neuronal cell membrane to increase intracellular levels of CuZnSOD protein . (A) Representative confocal microscopy images showing rhodamine fluorescence in CATH. A neurons following 1 or 3 hours of incubation with rhodamine-labeled free CuZnSOD protein or CuZnSOD nanozyme (400 U/mL). Magnification bar equals 10 μm. (B) Representative confocal microscopy images showing CuZnSOD immunofluorenscence staining in control CATH. A neurons and neurons treated (3 hours) with free CuZnSOD protein, PEG-SOD, or CuZnSOD nanozyme (400 U/mL). Magnification bar equals 10 μm. (C) Summary data (n=4 separate culture per group) of CuZnSOD immunofluorenscence in control CATH. a neurons (n=96 neurons) and neurons treated (3 hours) with free CuZnSOD protein (n=8 neurons), PEG-SOD (n=75 neurons) or CuZnSOD nanozyme (n=66 neurons). *p<0.05 vs. control and free CuZnSOD. a.u.=arbitrary units. Copyright permission from ref. 30.
3.2.2 In situ polymerization
Several of reportorial or functional protein molecules have been successfully polymerized in native site nanocapsules, include EGFP, HRP, BSA, Caspase-3 and SOD. Different from Physical adsorbed encapsulation, in situ polymerization-based encapsulation occurs on the surface of core materials, similar to interfacial polymerization. Du et al. utilized the “single protein nanocapsule” concept to encapsulate HRP and decorate the polymeric shell of nanocapsules with quantum dots (QDs) for a bioluminescence study [31]. The bioluminescence generated from HRP-mediated oxidation of luminol can well-overlap with the absorbance wavelengths of QDs, which enables their effective bioluminescence resonance energy transfer (BRET). The maximum BRET efficiency can be achieved by adjusting the enzyme/QD conjugation ratio. Though the SOD molecules have little ability of illumination, the in situ polymerization provides a perspective method to investigate the SOD molecular tracer based on the encapsulated QDs.
3.3 Magnetic nanoparticles
Magnetic nanoparticles (MNPs) have been extensively investigated in the field of biomedical applications, including drug delivery, chemical and biochemical separation and enrichment of trace amounts of specific targets. Particularly, magnetically-mediated delivery approaches can enhance the therapeutic profile by increasing localized concentration of target cargoes and minimizing non-specific interactions [32]. Muzykantov et al. investigated a biocompatible magnetic nanocarrierbased strategy for efficient encapsulation of two antioxidant enzymes, catalase and superoxide dismutase (SOD) [33]. The enzyme-loaded MNPs were assembled through hydrophobic and electrostatic interactions between iron oxide MNPs and proteins. The average size of enzyme-loaded MNPs was from 300 to 400 nm, with a protein loading efficiency of 20~33%. To demonstrate that antioxidant enzyme-loaded MNPs can rescue target cells from hydrogen peroxide toxicity, catalase-loaded MNPs were tested for intracellular delivery. Upon magnetic guidance, catalase-loaded MNPs with a negative net charge at pH 7.4 (-9.3±1.1 mv) were efficiently taken up by bovine aortic endothehal cells . A majority of cells that internalized catalase-loaded MNPs increase their resistance to oxidative stress and were rescued from hydrogen peroxide induced cell death. In the absence of magnetic field, only 10% cells were rescued (Fig. 8).
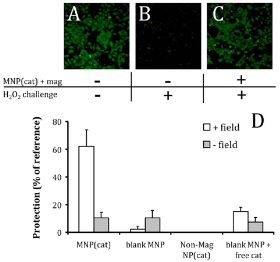
Fig. 8 Protection of HUVEC from oxidative stress through magnetic delivery of catalase loaded MNP. Viability of cells pretreated with MNP and exposed to 10 mM hydrogen peroxide for 5 hour was determined fluorimetrically after staining with Calcein AM. (A): Untreated cells used as reference. (B): Cells exposed to hydrogen peroxide only (no protection). (C): Cells treated under magnetic conditions with MNPencapsulated catalase. Original magnification ×100. (D): Quantification of the viability of hydrogen peroxide-challenged cells treated with catalase-loaded MNP in the presence of a high gradient magnetic field in comparison to controls. Copyright permission from ref. 33.
3.4 Protein-mediated carriers
In addition to polymer or magnetic nanocarriers, polypetides have also served as guides for transducing target proteins to the intracellular space of cells. Ming yang and his coworkers reported a novel intracellular delivery platform based on nanocapsules that consist of a single-protein core and thin polymer shell anchored covalently to the protein core [34]. In the experiment, the polymerizable vinyl groups were covalently linked to the protein in the first step and subsequently, the polymerization in an aqueous solution containing monomers and crosslinker results in each protein core being wrapped in a thin polymer shell. As illustrated in Fig. 9, this scheme enables the synthesis of protein nanocapsules with a non-degradable or degradable skin by using non-degradable or degradable corsslinkers, respectively. By appropriate choice of the cationic or neutral monomer, the surface charge of the nanocarrier and the release site of protein core can be controlled precisely. The protein cores can be chosen from a vast library of proteins, including enhanced green fluorescent protein (EGFP), horseradish peroxidase (HRP), bovine serum albumin (BSA), superoxide dismutase (SOD) and caspase-3 (CAS). In order to deliver more wide sorts of active proteins that could act on different cellular targets, a general mechanism for enzymatic degradation of the polymer nanocapsules and release of the protein cargo is needed. For this purpose, Anuradha Biswas and her coworkers designed the polymer nanocapsule that can disintegrate and release proteins in response to the essential endoprotease furin, which is a ubiquitous proprotein convertase that could cleavage of the papillomavirus minor capsid protein L2 for necessary dissociation of the capsid, release of viral DNA, and subsequent transfections [35]. At first, they made the noncovalent encapsulate the target protein cargo in a thin, positively charged polymer layer, using two monomers and furin-cleavable peptides as cross-linkers (CLs). Then, they utilized differential acid-labile protection groups on the side chains of amino acids to synthesize the CL, allowed acryolation of only the N- and C-terminal free amine groups while preserving the arginine groups unmodified for furin recognition. Additionally, complete synthesis on amide resin resulted in high yield and purity of the final peptide product. Using this established method, they chose eGFP as signal indication and build the NLS-eGFP nano protein cargo. After transferred the NLS-eGFP into different Chinese hamster ovary (CHO) cell lines, including FD11 (furindeficient strain) and FD11+ (FD11 strain transfected with an overexpressed furin gene) and eGFP fluorescence was observed and compared between FD11 and FD11+ strains. The results showed that the fluorescence in FD11+ strain is 1000 higher than that in DF11 strain. This experiment demonstrated that the NLS-eGFP nano protein cargo was successfully transferred and released into nuclear site after the nanocapsule was degraded.
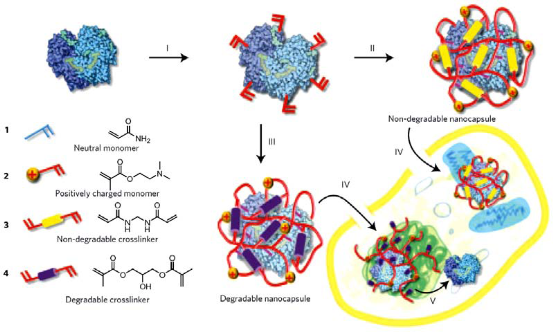
Fig. 9 Schematic showing the synthesis and cellular uptake of cationic single-protein nanocapsules with degradable and non-degradable polymeric shells prepared by in situ co-polymerization of acrylamide, 3-dimethylaminoethyl methacrylate and non-degradable crosslinker methylenebisacrylamide or acid-degradable dimethacrylate. (I):formation of polymerizable proteins by conjugation polymerizable acryl groups to the protein surface. (II): formation of non-degradable nanocapsules from 1, 2 and 3. (III): formation of degradable nanocapsules from 1, 2 and 4. (IV): cellular uptake of the degradable or non-degradable nanocapsules via endocytosis. (V): shells of degradable nanocapsules break down after internalization to release the protein cargoes, allowing them to interact with large molecular substrates. Copyright permission from ref. 34.
4. The factors affecting the delivery efficiency of SOD nanocarriers
As a primary antioxidant enzyme inhibiting oxidative stress and the nanocarriers of SODs molecules have several type and structures. Whereas several factors that affect the delivery efficiency of the SOD nanocarriers should be reminded in animal studies or clinical trails.
4.1 Delivery vehicle
Antioxidant enzymes have been formulated into nonpolymeric nanocarriers including traditional selfassembly phospholipid carriers, liposomes and magnetic nanocarriers or diverse pplyplex complexes as mentioned above. For example, SOD associated with the surface of liposomes was shown to be more resistant than free SOD to inactivation by high concentrations of H2O2 [36]. The early work of liposome based SOD delivery to cultured endothelial cells carried by Freeman et al., indicated a 6~12 folds increase in SOD activity compared with the control cells treated with free SOD and the cells received liposomal SOD were more resistant to oxidative damage by hyperoxial [37]. Another experiment of anti-inflammatory effect of acetylated hydrophobic derivative of SOD in a rat arthritic model showed an improved loading efficiency and bioactivity in target site. The change in conformation reduced the effect of release rate on the activity of the liposome, prolonged circulation time and increased activity of the enzymosome [38]. The circulation time of the PEGylated liposomes increased regardless of the SOD type included and a faster anti-inflammatory effect was observed with the As-SOD PEG liposome versus the plain SOD PGE lioosomes [39]. Batrakova et al. developed a macrophage-driven system for the delivery of catalase to the brain. The catalase was elecrostatically complexed with a cationic block copolymer, polyethylene-imine-poly (ethylene glycol) (PEI-PEG). The PEI-PEG nanozyme shielding protected the enzyme within the structure were phagocytized by macrophages, targeted by subsequent migration of the cell to the inflamed brain in a mouse model of Parkinson’s disease [40]. The similar results of the formed SOD nanozymes alleviated neuronal oxidative stress after local administration in the CNS in rodents showed the promising therapeutic efficacy of this delivery system [30]. Although the intracellular transport mechanism of PEG-containing pluronics of SOD remains vague, the SOD-pluronic conjugates were reported to deliver enzymatically active SOD to neuronal cells more effective cell uptake ratio than naked SOD or PEG-SOD without neuronal toxicity in cultured cells and in vivo [41].
4.2 Loading capacity and nanocarrier volume
The functions of nanocarriers include protection of antioxidant cargoes from inactivation, improved vascular targeting and intracellular delivery. In order to achieve the maximum effect of antioxidant, the nanocarriers should be designed for maximum loading efficacy of SOD molecules. However, the larger carriers may cause side effects and exceed size limits for adequate circulation. Furthermore, the overloading of nanocarriers may result in limited control of the drug release profile, bigger volume of the cargoes may lead to the consequences of circulation and tissue uptake [42,43]. By using a single-step solvent extraction method, the poly-nanocarriers (such trolox ester and SOD) can be formulated into nanoparticles of size 100~250 nm, that are able to quench ROS and suppress oxidative stress in cell culture of endothelial cells exposed to pro-oxidant challenge [44]. Another interesting experiment is introduce the PEG400 as a polymer stabilizer, these particles could achieve ca. 10~15 μm and an over 50 days active durative [45]. However, their size exceeded the circulation limit of less than 500nm in diameter and they are often been cleared rapidly due to retention in the microvasculature [46]. At the same time, the small (<20 nm) volume of nanocarriers usually extravasated via vascular pores and retained in parenchymal cells. This will lead to a controllable AOEs cargo on principle of tissue or vascular target. Recently, several proteins including SOD were covalently funcionalized with vinyl groups follow by free radical polymerization in the presence of other diacrylate monomers, which resulted in encapsulation of protein molecule in a nanometer thick (0~5 nm) polymer shell [34]. Even though the utilities for delivery of AOEs has not been fully studied, polymer nanocapsues incorporating disulfide bridges that can degrade in a reducing environment could be applicable for release of the cargo triggered in the host cell [47].
4.3 Geometry of nanocarriers for antioxidants
Spherical shape is the most common among the polymeric nanocarriers (PNCs) formulation that range from relatively small sphere (50~300 nm) [48] to large spheroid (300~1000 nm) [49] with diverse outline, including polymersomes, dendrimers and polyplexes. Recently, various nonspherical nanocarriers such as carbon nanotubes have been yielded via chemical or physical reaction, and have been recognized that the geometry of nanocarrier could modulates drug delivery functions. Another class of nonspherical model PNCs includes flat elliptical disks [50,51] with format of emulsions or micron-scale particles [52]. Furthermore, depending on the molar ratio of hydrophilic PEG or polyethylene oxide (PEO) block to the hydrophobic block, filomicelles nanocarriers have been made for antioxidant delivery [53]. Filomicelles are long tubular structures with width ca. 10~40 nm, length ≤50 μm and ca. 42~50% PEG hydrophilic surfaces [54]. The filomicelles degrade experiments showed a small water diffusion distance throughout it, the cross sectional radii <20 nm [55]. Since PCL has a rapid degradation character and PEO does not degrade in water, a well controlled release system of nanocarriers could be made via manipulate the ratio of PEO:PCL in the filomicelles synthesis process. Comparison spherical carriers of intermediate size circulate for many hours and even days, the highly flexible filomicelles have circulation t1/2 approaching 1 week in mice [56]. The longer filomicelles circulate longer and are not readily internalized by macrophages under flow with less vascular collisions, extravasation and phagocytosis. The carrier’s flow-aligning structure provides extensive drag forces from directional flow that oppose phagocytosis [50].
4.4 The core protection of the nanocarriers
As an alternative approach, one can enhance enzyme stability through the addition of a protecting molecule. Since inactivation occurs in part due to the enzyme denaturation in the hydrophobic/hydrophilic interface, the addition decoy protein (such as serum albumin) can prolong the activity of the loaded enzyme at the expense of total loading capacity. Since the cargo surface is not attached to protein specifically, by allowing a nonactive protein to absorb to the surface instead, the active antioxidant enzyme is covered in the core site and thereby able to maintain its activity for a relative prolonged half life. For instance, the activity of SOD released from PLGA nanoparticles along with protective albumin retained activity after 7 days, versus 4 days in the case of omitting albumin [57]. Recently, several AOEs including SOD were covalently functionalized with vinyl groups followed by free radical polymerization in the presence of other diacrylate monomers, which produce an encapsulation of protein molecule in a nanometer thick polymer shell [34]. The polymer properties and degradation rate were controlled by monomer selection; the encapsulation of AOEs in the polymer shell can provide protection from proteolytic inactivation, thereby extending therapeutic duration of SOD molecule [49].
4.5 Target and carrier degradation
Systemic medication and local treatment are common measures in clinical iatrology, these mechanisms of delivery are not specific and provide no targeting to select the cells or organs suffering oxidative stress. In this context, targeted delivery of antioxidants to specific cells is a primary therapeutic target and eliminate local oxidative stress represents an important and challenging goal [58]. Specific conjugation with antibodies to ICAM and PECAM provides intracellular target delivery of multivalent conjugate and nanocarriers. This approach providing the AOEs molecule potential ‘stealth’ properties and can be conjugated with multi-antibodies to the nanocarriers during its formulations. For instance, anti-PECAM/coated PNC loaded with catalase providing delivery of 12% of the injected dose in the lungs as compared with 2% for nontargeted control nanocarriers. The anti-PECAM/coated catalase inhibited the ROS leves in the lungs obviously than native nanocarriers [59]. Biodegradability is the main requirements for biocompatibility of AOEs delivery system if its size not permits excretion via physiological pathways. Two typical degradable material with different degradation kinetics are polyanhydrides and polyesters. The former process a degradatdion time from hours to weeks that could achieved by alteration of the ratio of constituent copolyers [60], but the later degradation time ranges from weeks to years [61]. Spherical polyanhydrides carriers have been synthesized on the nanoscale [62], whereas, how to control its shape and decrease its degradation kinetics have not been fully documented. The rapid release kinetics restricts polyanhydrides application in AOEs nanocarriers synthesis unless it been used in local and trivial inflammation. In contrast, nanometer scale polyester spheres provide uniform water degradation and erosion, its internal degradation can actually equalize exterior degradation and lead to an efficient diffusion inside the carrier [63]. This made polyester a promising material in nanocarrier synthesis of AOEs. PEO-PLA polymersomes and PEO-PCL filomicelles degrade primarily by PLA and PCL hydrolysis [61,64], result in pore-preferring copolymers [65] and filomicelle fragmentation [66]. The degradation kinetics could be controlled by blending in nondegradable diblocks, such as PEO and PEE that excreted via renal pathway. In this context, the polymersomes synthesized in different ratio of PEO-PCL or PEO-PLA was widely used in nanocarries of AOEs. Environmental pH plays a role in polymersomes degradation. Degradable polymers typically contain hydrolysable bonds and undergo faster acid-catalyzed hydrolysis at low pH [67]. The latest investigation demonstrated that the rate of polymersome hydrolysis and release of loaded drugs is faster at acidic lysosomal pH (5.0) than any other organelles with neuter pH (7.2~7.4) [68]. Interestingly, polyanhydrides degrade more slowly at acidic pH with far more rapid degradation in basic solutions [69,70]. Take into account pH difference in diverse organelles of target cell, various pH sensitive nanocarries could be synthesized in a near future.
5. Challenges and prospects
Although SOD exhibits great application prospects in clinical anti-oxidization therapy, SOD based novel pharmaceutical development still existed many difficulties. For example, how to highly efficiently deliver SOD into therapeutic target locus in vivo, how to make endosome or endolysomes filled with SOD effectively release SOD into cytoplasm. Nanomaterials can induce cells produce lot of oxidative radicals, how to use SOD to resist oxidative radicals caeused by nanomaterials, all these problems have become great challenges. Delivering functional proteins to the intracellular and interact with special targets of cells, is a highly promising therapeutic application. Especially for such antioxidant molecule as SOD and Coenzyme Q10 have been modified with nanotechnology and transferred into intracellular, the promising biotechnology will exert a significant function in removing wrinkles, rebuilding tissue, and extending the length of human life. Despite those efforts, active or functional proteins, such as SOD etc., intracellular delivery is still in its initial stage, and further research is needed to fully reveal its vast potential. Furthermore, the relationship between function and structure should be taken into account when constructing nanomaterial platforms. Meanwhile, the delivery characteristics of these nanocarriers were investigated with cell lines in vitro, the mechanism of its biological feedback and the interaction in vivo should be further explored. Nowadays, many difficulties still need to be overcome to obtain the nanocarriers with effective and efficient properties. For example, covalent modifications may lead to the function loss to the delivery protein and noncovalent strategies are often producing the weak stable products in serum. Efficient intracellular transport is still a challenge as many nanocarriers can not escape the cytosol from the endosomal pathway efficiently [71]. How to control the release of cargoes through a timed mechanism instead of a burst release is another desired function of nanocarriers. This can be implemented with synthesis of stimuli-responsive nanocarriers to get the release in an “on demand” way. It is worthy of further development of the “smart” nanocarriers by contriving materials with sensitive responses to such environmental conditions as pH, redox, endosomal enzyme activity, temperature and physical signals applied externally such as magnetic field and light [72]. At last, the terminal delivery goals are the targeting of specific cells or organs for disease-specific therapies. In vivo, the protein nanocarriers may encounter numerous obstacles en route to their end-point. In this process, the most important is to allow these nanocarriers to accumulate in the designed position, by the enhanced permeability and retention (EPR) effect mediated-passive targeting or active targeting through conjugating targeting moieties, such as antibodies, receptor ligands (peptides, vitamins, and carbohydrates) and aptamers [25]. The effects of these nanocarrier systems can be evaluated by the experiential knowledge accumulated previously of nanomedicine, including biocompatibility, stability, uptake and biological interactions [73].
6. Concluding Remarks
In conclusion, many advances discussed in this review have been used to account these critical issues, including vehicle tailoring and protein engineering. We imagine that crucial progress will be made and new generations of protein nanocarriers will be developed in a near future. Modification and encapsulation antioxidant proteins, especially for SOD and coenzyme Q that closely related with mammals diseases, prolong human’s life, will bring a promising application within the interdisciplinary collaborated research [30].
Acknowledgements
This work was supported by Chinese 973 Project (2010CB933901), the National Natural Science Foundation of China (No.81225010, 81101169 and 31100717), New Century Excellent Talent of Ministry of Education of China (NCET-08-0350), Special Infection Diseases Key Project of China (2009ZX10004-311) and Doctorial Position Budget (20070248050).
References
1. Borm P.J., Robbins D., Haubold S., Kuhlbusch T., Fissan H., Donaldson K. The potential risks of nanomaterials: a review carried out for ECETOC. Part Fibre Toxicol, 2006; 3: 1-35.
2. Keilin T.M.D. Haemocuprein and hepatocuperin, copper-protein compounds of blood and liver in mamals. Series B. Biolog. Sci., 1938; 126(9): 303-315.
3. Hartree D.K.E.F. Cytochrome oxidase and the 'Pasteur enzyme'. Nature, 1953; 171: 413-416.
4. McCord J.M., Fridovich I. The reduction of cytochrome c by milk xanthine oxidase. J. Biol. Chem. 1968; 243: 5753-5760.
5. Shimoda-matsubayashi S., Matsumine H., Kobayashi T., Nakagawahattori Y., Shimizu Y., Mizuno Y. Structural Dimorphism in the Mitochondrial Targeting Sequence in the Human Manganese Superoxide Dismutase Gene. Biochem. Biophys. Res. Commun. 1996; 226: 561-565.
6. Youn H.D., Kim E.J., Roe J.H., Hah Y.C., Kang S.O. A novel nickel-containing superoxide dismutase from Streptomyces spp. Biochem. J. 1996; 318: 889-896.
7. Cocco D., Calabrese L., Rigo A., Marmocchi F., Rotilio G., Biologica C. Preparation of selectively metal-free and metal-substituted derivatives by reaction of Cu-Zn superoxide dismutase with diethyldithiocarbamate. Biochem. J. 1981; 199: 675-680.
8. Lamb A.L., Torres A.S., Halloran T.V.O., Rosenzweig A.C. Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat. Struct. Biol., 2001; 8:751-755.
9. Banci L., Benedetto M., Bertini I., Del Conte R., Piccioli M., Viezzoli M.S. Solution structure of reduced monomeric Q133M2 copper, zinc superoxide dismutase (SOD). Why is SOD a dimeric enzyme? Biochemistry, 1998; 37:11780-11791.
10. Zelko I.N., Mariant T.J., Folz R.J. Superoxide dismutase multigene family: A comparison of the CuZnSOD (SOD1), MnSOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radical Biol. Med. 2002; 33:337-349.
11. Hough M., Hasnain S.S. Crystallographic structures of bovine copper-Zn superoxide dismutase reveal asymmetry in two subunits: functionally important three and five coordinate copper site captured in the same crystal. J. Mol. Biol., 1999; 287:579-592.
12. Hough M., Hasnain S.S. Structure of Fully Reduced Bovine Copper Zinc Superoxide Dismutase at 1.15 Å. Structure, 2003; 11:937-946.
13. Fink R.C., Scandalios J.G. Molecular evolution and structure--function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys., 2002; 399:19-36.
14. Borgstahl G.E.O, Parge H.E., Hickey M.J., Beyer W.F., Hallewell R.A., Tainer J.A. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell, 1992; 71:107-118.
15. Edwards R.A., Baker H.M., Whittaker M.M., Whittaker J.W., Jameson G.B., Baker E.N. Crystal structure of Escherichia coli manganese superoxide dismutase at 2.1-Å resolution. J Biol Inorg Chem. 1998; 3: 161-171.
16. Carlioz A., Ludwig M.L., Stallings W.C., Fee J., Steinman H.M., Touati D. Iron superoxide dismutase. Nucleotide sequence of the gene from Escherichia coli K12 and correlations with crystal structures. J. Biol. Chem. 1988; 263:1555-1562.
17. Huber R., Wilharm T., Huber D., Trincone A., Burggraf S., König H. Aquifex pyrophilus gen. nov. sp. nov., Represents a Novel Group of Marine Hyperthermophilic Hydrogen-Oxidizing Bacteria. Syst. Appl. Microbiol., 1992; 15:340-351.
18. Lim J.H., Yu Y.G., Choi I.G., Ryu J.R., Ahn B.Y., Kim S.H. Cloning and expression of superoxide dismutase from Aquifex pyrophilus, a hyperthermophilic bacterium. Febs. Lett., 1997; 406:142-146.
19. Lim J.H., Yu Y.G., Han Y.S., Cho S., Ahn B.Y., Kim S.H.. The crystal structure of an Fe-superoxide dismutase from the hyperthermophile Aquifex pyrophilus at 1.9 A resolution: structural basis for thermostability. J. Mol. Biol., 1997; 270:259-274.
20. Patel L.N., Zaro J.L., Shen W.C. Cell penetrating peptides: intracellular pathways and pharmaceutical perspectives. Pharmaceut. Res., 2007; 24:1977-1992.
21. Gu Z., Biswas A., Zhao M., Tang Y. Tailoring nanocarriers for intracellular protein delivery. Chem. Soc. Rev., 2011; 40:3638-55.
22. Petri W. Protein transduction domains: are they delivering? Trends Pharmacol. Sci., 2003; 24:210-212.
23. Gao J., Xu B. Applications of nanomaterials inside cells. Nano Today, 2009; 4:37-51.
24. Lee K.Y., Yuk S.H. Polymeric protein delivery systems. Prog. Polym. Sci., 2007; 32:669-697.
25. Peer D., Karp J.M., Hong S., Farokhzad O.C., Margalit R., Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotech. 2007; 2:751-760.
26. Chou L.Y.T., Ming K., Chan W.C.W. Strategies for the intracellular delivery of nanoparticles. Chem. Soc. Rev., 2011; 40:233-245.
27. Wang J., Hu T., Liu Y., Zhang G., Ma G., Su Z. Kinetic and stoichiometric analysis of the modification process for N-terminal PEGylation of staphylokinase. Anal. Biochem., 2011; 412:114-116.
28. Solaro R. Targeted delivery of proteins by nanosized carriers. J. Polym. Sci., Part A: Polym. Chem. 2008; 46:1-11.
29. Liguori L., Marques B., Villegas-Mendez A., Rothe R., Lenormand J.L. Liposomes-mediated delivery of pro-apoptotic therapeutic membrane proteins. J. Controlled Release, 2008; 126:217-227.
30. Rosenbaugh E.G., Roat J., Gao L., Yang R.F., Manickam D.S., Yin J.X. The Attenuation of Central Angiotensin II-dependent Pressor Response and Intra-neuronal Signaling by Intracarotid Injection of Nanoformulated Copper/Zinc Superoxide Dismutase. Biomaterials, 2011; 31:5218-5226.
31. Du J., Yu C., Pan D., Li J., Chen W., Yan M. Quantum-dot-decorated robust transductable bioluminescent nanocapsules. J. Am. Chem. Soc.,2010; 132:12780-12781.
32. Gao J., Gu H., Xu B. Multifunctional magnetic nanoparticles: design, synthesis, and biomedical applications. Accounts. Chem. Res., 2009; 42:1097-1107.
33. Michael C., Elizabeth H., Levy R.J., Muzykantov. Endothelial delivery of antioxidant enzymes loaded into nonpolymeric magnetic nanoparticles. Biochemistry, 2010; 29:8885-8893.
34. Yan M., Du J., Gu Z., Liang M., Hu Y., Zhang W. A novel intracellular protein delivery platform based on single-protein nanocapsules. Nature nanotech. 2010; 5:48-53.
35. Biswas A., Joo K.I., Liu J., Zhao M., Fan G., Wang P. Endoproteasemediated intracellular protein delivery using nanocapsules. ACS nano, 2011; 5:1385-1394.
36. Nagami H., Yoshimoto N., Umakoshi H., Shimanouchi T., Kuboi R. Liposome-assisted activity of superoxide dismutase under oxidative stress. J. Biosci. Bioeng., 2005; 99:423-428.
37. Freeman B., Young S.L., Crapo J.D. Liposome-mediated augmentation of superoxide dismutase in endothelial cells prevents oxygen injury. J. Biol. Chem. 1983; 258:12534-12542.
38. Gaspar M.M., Martins M.B., Corvo M.L., Cruz M.E.M. Design and characterization of enzymosomes with surface-exposed superoxide dismutase. Biochim. Biophys. Acta, 2003; 1609:211-217.
39. Gaspar M.M., Boerman O.C., Laverman P., Corvo M.L., Storm G., Cruz M.E.M. Enzymosomes with surface-exposed superoxide dismutase: in vivo behaviour and therapeutic activity in a model of adjuvant arthritis. J. controlled release, 2007; 117:186-195.
40. Batrakova E.V., Li S., Reynolds A.D., Mosley R.L., Tatiana K., Kabanov A.V. A macrophage-nanozyme delivery system for Parkinson’s disease. Bioconjugate Chem., 2009; 18:1498-1506.
41. Yi X, Zimmerman M.C., Yang R.F., Tong J., Vinogradov S.V., Kabanov A.V. Pluronic-Modified Superoxide Dismutase 1 (SOD1) Attenuates Angiotensin II-Induced Increase in Intracellular Superoxide in Neurons. Free. Radical. Biol. Med., 2010; 49:2-2.
42. Leonarduzzi G., Testa G., Sottero B., Gamba P., Poli G. Design and development of nanovehicle-based delivery systems for preventive or therapeutic supplementation with flavonoids. Curr. Med. Chem., 2010; 17:74-95.
43. Kumar V., Hong S.Y., Maciag A.E., Saavedra J.E., Douglas H., Prud R.K. Stabilization of the nitric oxide (NO) prodrugs and anticancer leads, PABA/NO and double JS-K, through incorporation into PEG-protected nanoparticles. Mol. Pharm., 2011; 7:291-313.
44. Wattamwar P.P., Mo Y., Wan R., Palli R., Zhang Q., Dziubla T.D. Antioxidant Activity of Degradable Polymer Poly(trolox ester) to Suppress Oxidative Stress Injury in the Cells. Adv. Funct. Mater., 2010; 20:147-154.
45. Giovagnoli S., Luca G., Casaburi I., Blasi P., Macchiarulo G., Ricci M., Long-term delivery of superoxide dismutase and catalase entrapped in poly(lactide-co-glycolide) microspheres: in vitro effects on isolated neonatal porcine pancreatic cell clusters. J. controlled release, 2005; 107:65-77.
46. Fiore V.F., Lofton M.C., Roser-Page S., Yang S.C., Roman J., Murthy N. Polyketal microparticles for therapeutic delivery to the lung. Biomaterials, 2010; 31:810-817.
47. Kim E., Kim D., Jung H., Lee J., Paul S., Selvapalam N. Facile, template-free synthesis of stimuli-responsive polymer nanocapsules for targeted drug delivery. Angew. Chem. Int. Ed., 2010; 49: 4405-4408.
48. Saad M., Garbuzenko O.B., Ber E., Chandna P., Khandare J.J., Pozharov V.P. Receptor targeted polymers, dendrimers, liposomes: which nanocarrier is the most efficient for tumor-specific treatment and imaging? J. Controlled Release, 2008; 130:107-114.
49. Dziubla T.D., Karim A., Muzykantov V.R. Polymer nanocarriers protecting active enzyme cargo against proteolysis. J. Controlled Release, 2005; 102:427-439.
50. Champion J.A., Mitragotri S. Role of target geometry in phagocytosis. PNAS, 2006; 103:4930-4934.
51. Muro S., Garnacho C., Champion J.A., Leferovich J., Schuchman E.H., Mitragotri S. Control of Endothelial Targeting and Intracellular Delivery of Therapeutic Enzymes by Modulating the Size and Shape of ICAM-1-targeted Carriers. Mol. Ther., 2008; 16:1450-1458.
52. Xu S., Nie Z., Seo M., Lewis P., Kumacheva E., Stone H. Generation of monodisperse particles by using microfluidics: control over size, shape and composition. Angew. Chemie. Int. Ed., 2005; 44:724-728.
53. Discher D.E., Eisenberg A. Polymer vesicles. Science, 2002; 297:967-973.
54. Dalhaimer P., Bates F.S., Discher D.E. Single Molecule Visualization of Stable , Stiffness-Tunable , Flow-Conforming Worm Micelles. Macromolecules, 2003; 36:6873-6877.
55. Simone E.A., Dziubla T.D., Colon-gonzalez F., Discher D.E., Muzykantov V.R. Effect of Polymer Amphiphilicity on Loading of a Therapeutic Nanocarriers. Biomacromol., 2007; 8:3914-3921.
56. Geng Y., Dalhaimer P., Cai S., Tsai R., Tewari M., Minko T. Shape effects of filaments versus spherical particles in flow and drug delivery. Nature nanotech. 2007; 2:249-255.
57. Reddy M.K., Wu L., Kou W., Ghorpade A., Labhasetwar V. Superoxide Dismutase-Loaded PLGA Nanoparticles Protect Cultured Human Neurons Under Oxidative Stress. Appl. Biochem. Biotech., 2008; 29:565-577.
58. Muro S., Dziubla T., Qiu W., Leferovich J., Cui X., Berk E. Endothelial Targeting of High-Affinity Multivalent Polymer Nanocarriers Directed to Intercellular Adhesion Molecule 1. Pharmacology, 2006; 317:1161-1169.
59. Dziubla T.D., Shuvaev V.V., Hong N.K., Hawkins B., Takano H., Simone E. Endothelial targeting of semi-permeable polymer nanocarriers for enzyme therapies. Biomaterials, 2009; 29:215-227.
60. Tabata Y., Gutta S., Langer R. Controlled delivery systems for proteins using polyanhydride microspheres. Pharmaceut. Res., 1993; 10:487-496.
61. Siepmann J. Göpferich A. Mathematical modeling of bioerodible, polymeric drug delivery systems. Adv. Drug Delivery Rev., 2001; 48:229-247.
62. Pfeifer B., Burdick J., Langer R. Formulation and surface modification of poly(ester-anhydride) micro- and nanospheres. Biomaterials, 2005; 26:117-124.
63. Schliecker G., Schmidt C., Fuchs S., Kissel T. Characterization of a homologous series of d, l-lactic acid oligomers; a mechanistic study on the degradation kinetics in vitro. Biomaterials, 2003; 24: 3835-3844.
64. Shive M., Anderson J. Biodegradation and biocompatibility of PLA and PLGA microspheres. Adv. Drug Delivery Rev., 1997; 28:5-24.
65. Ahmed F., Discher D.E. Self-porating polymersomes of PEG-PLA and PEG-PCL: hydrolysis-triggered controlled release vesicles. J. controlled release, 2004; 96:37-53.
66. Geng Y., Discher D.E. Hydrolytic degradation of poly (ethylene oxide)-block-polycaprolactone worm micelles. J. Am. Chem. Soc., 2005; 127:12780-12781.
67. Shih C. A graphical method for the determination of the mode of hydrolysis of biodegradable polymers. Pharmaceut. Res., 1995; 12:2036-2040.
68. Ahmed F., Pakunlu R.I., Srinivas G., Brannan A., Bates F., Klein M.L. Shrinkage of a rapidly growing tumor by drug-loaded polymersomes: pH-triggered release through copolymer degradation. Mol. Pharm., 2006; 3:340-350.
69. Muzykantov V.R., Atochina E.N., Ischiropoulos H., Danilovt S.M., Fisher A.B. Immunotargeting of antioxidant enzymes to the pulmonary endothelium. PNAS, 1996; 93:5213-5218.
70. Göpferich A. Mechanisms of polymer degradation and erosion. Biomaterials, 1996; 17:103-114.
71. Palm T., Esfandiary R., Gandhi R. The effect of PEGylation on the stability of small therapeutic proteins. Pharm. Dev. Technol., 2011; 16:441-448.
72. Stuart M.C., Huck W.T.S., Genzer J., Müller M., Ober C., Stamm M. Emerging applications of stimuli-responsive polymer materials. Nature Mater., 2010; 9:101-113.
73. Adiseshaiah P.P., Hall J.B., McNeil S.E. Nanomaterial standards for efficacy and toxicity assessment. WIRES Nanomed. Nanobi., 2009; 2:99-512.
Copyright:(c) 2012 C. Li et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.